visualize marker genes LECs
Load packages
## load packages
suppressPackageStartupMessages({
library(dplyr)
library(reshape2)
library(ggplot2)
library(cowplot)
library(purrr)
library(Seurat)
library(tidyverse)
library(ggpubr)
library(runSeurat3)
library(here)
library(ggsci)
library(pheatmap)
library(scater)
})load seurat object
basedir <- here()
seurat <- readRDS(file= paste0(basedir,
"/data/AllPatWithoutCM_LEConly_intOrig_seurat.rds"))
DefaultAssay(object = seurat) <- "RNA"
seurat$intCluster <- seurat$integrated_snn_res.0.25
Idents(seurat) <- seurat$intCluster
## set col palettes
colPal <- colPal <- c(pal_nejm()(8))[1:length(unique(seurat$intCluster))]
names(colPal) <- unique(seurat$intCluster)
colLab <- c("#79AF97FF","#374E55FF","#B24745FF","#DF8F44FF","#6A6599FF",
"#D595A7FF")
names(colLab) <- c("SCScLEC","MedCapsLEC", "SCSfLEC", "MedSinusLEC",
"ParacortLEC","ValveLEC")
colPat <- c(pal_nejm()(7),pal_futurama()(12))[1:length(unique(seurat$patient))]
names(colPat) <- unique(seurat$patient)
colCond <- c("#6692a3","#971c1c","#d17d67")
names(colCond) <- unique(seurat$cond)
colGrp <- pal_uchicago()(length(unique(seurat$grp)))
names(colGrp) <- unique(seurat$grp)
colOri <- pal_npg()(length(unique(seurat$origin)))
names(colOri) <- unique(seurat$origin)
colCond2 <- c("#6692a3","#971c1c")
names(colCond2) <- c("resting", "activated")visualize data
clustering
## visualize input data
DimPlot(seurat, reduction = "umap", cols=colPal)+
theme_bw() +
theme(axis.text = element_blank(), axis.ticks = element_blank(),
panel.grid.minor = element_blank()) +
xlab("UMAP1") +
ylab("UMAP2")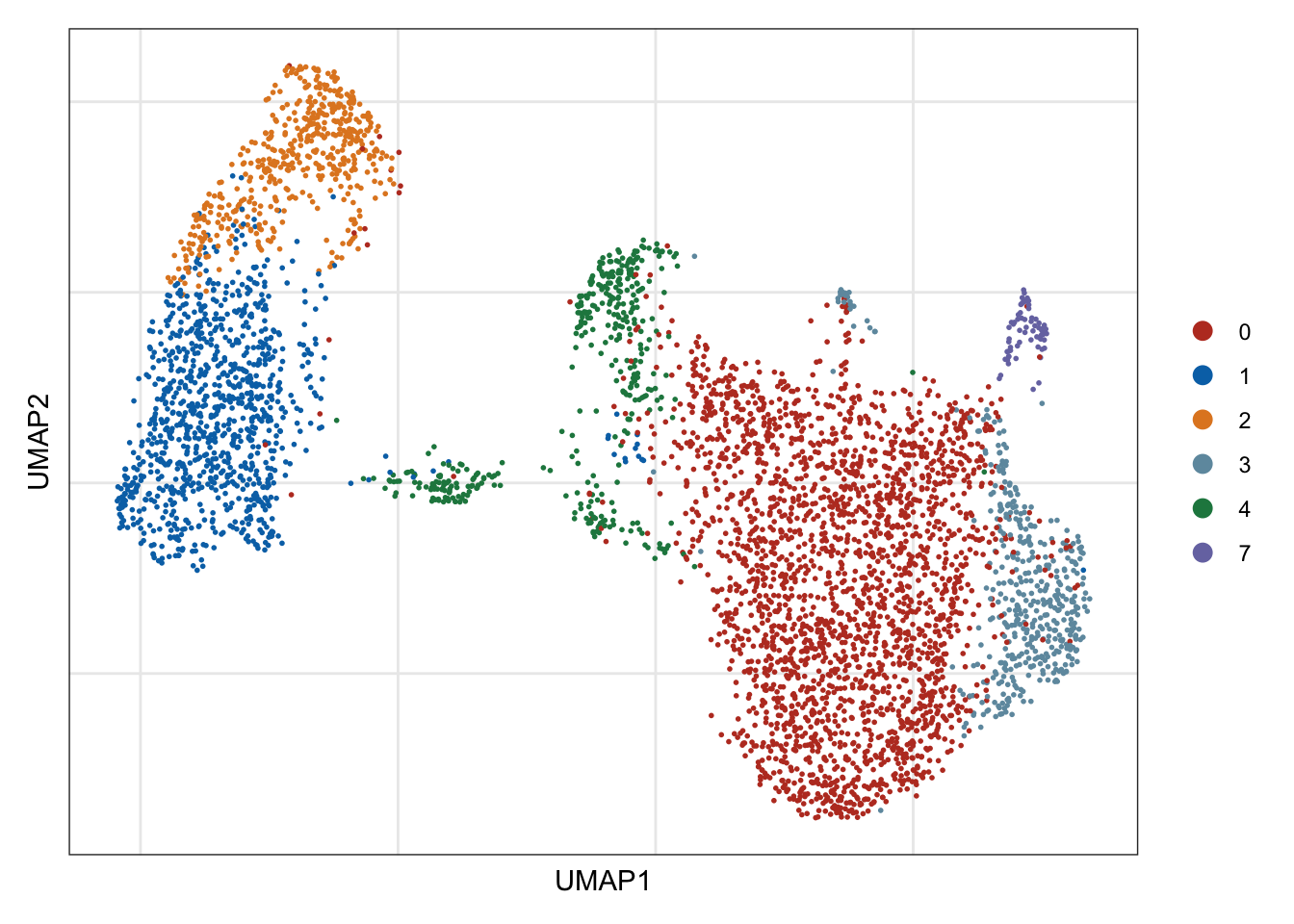
DimPlot(seurat, reduction = "umap", cols=colPal, pt.size=0.5)+
theme_void()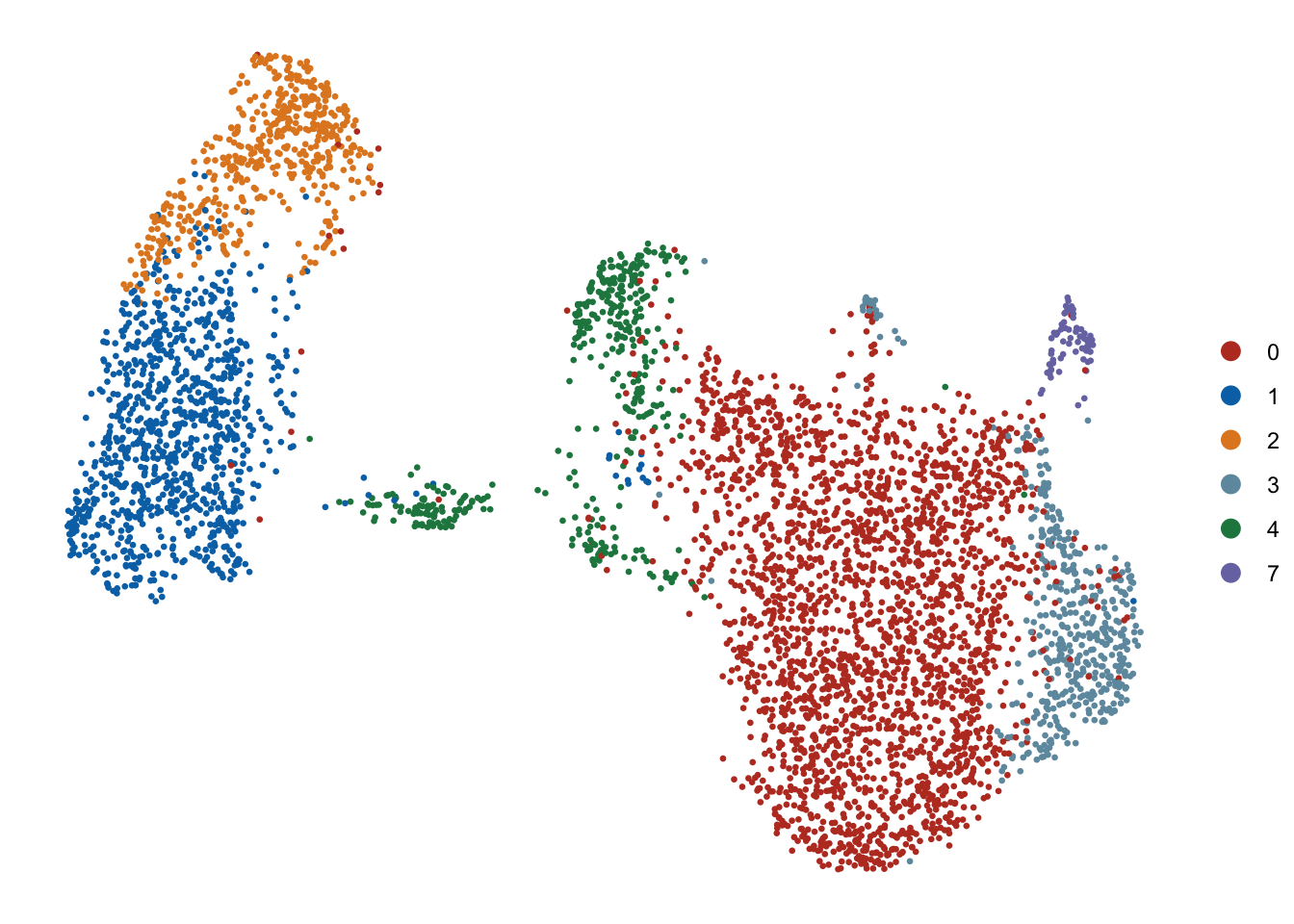
cluster split by cond
DimPlot(seurat, reduction = "umap", cols=colPal, pt.size=0.5,
split.by = "cond2")+
theme_void()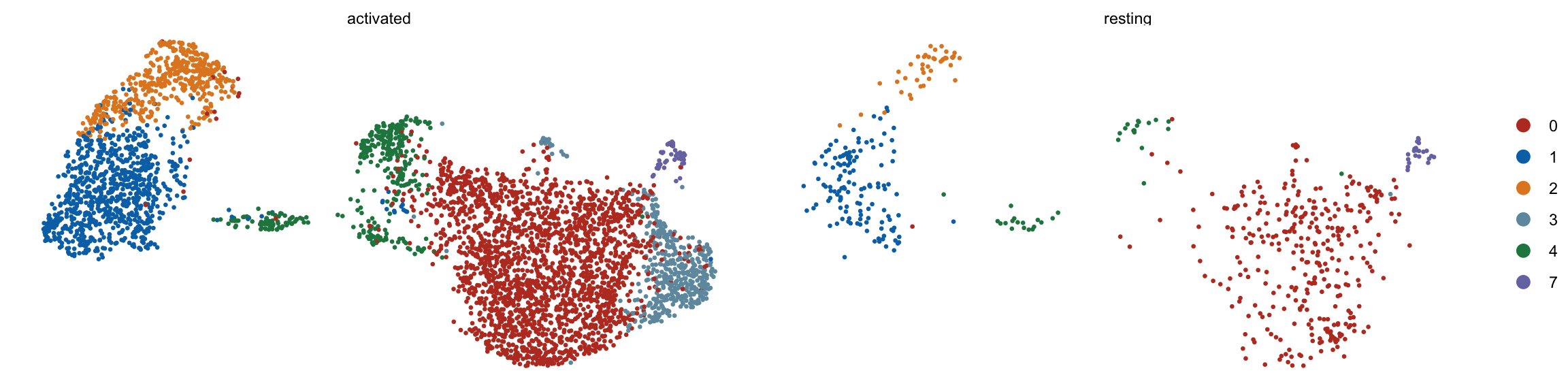
patient
## visualize input data
DimPlot(seurat, reduction = "umap", cols=colPat, group.by = "patient")+
theme_bw() +
theme(axis.text = element_blank(), axis.ticks = element_blank(),
panel.grid.minor = element_blank()) +
xlab("UMAP1") +
ylab("UMAP2")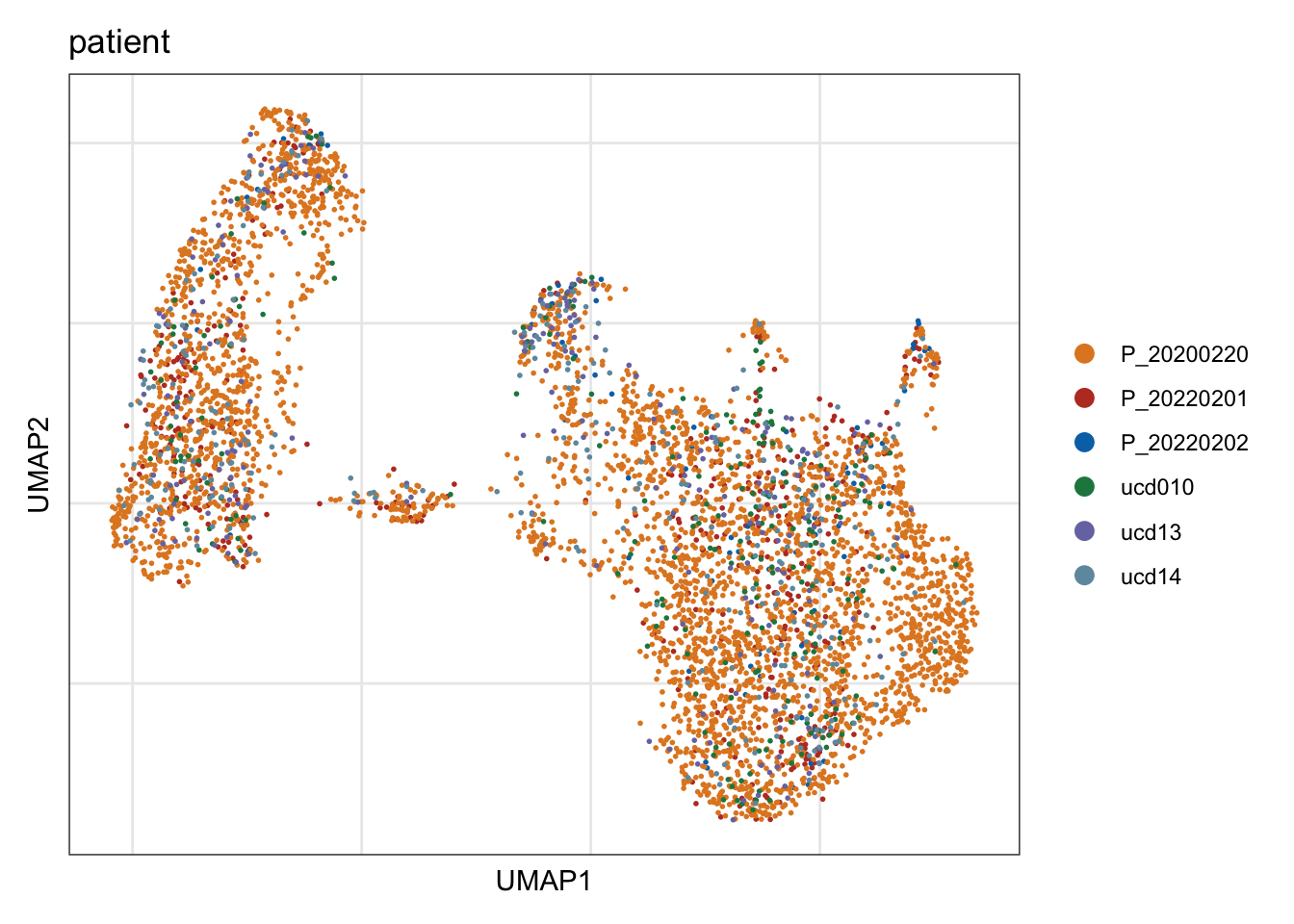
DimPlot(seurat, reduction = "umap", cols=colPat, group.by = "patient",
pt.size=0.5, shuffle = T)+
theme_void()
cond
## visualize input data
DimPlot(seurat, reduction = "umap", cols=colCond2, group.by = "cond2")+
theme_bw() +
theme(axis.text = element_blank(), axis.ticks = element_blank(),
panel.grid.minor = element_blank()) +
xlab("UMAP1") +
ylab("UMAP2")
DimPlot(seurat, reduction = "umap", cols=colCond2, group.by = "cond2",
pt.size=0.5, shuffle = T)+
theme_void()
grp
## visualize input data
DimPlot(seurat, reduction = "umap", cols=colGrp, group.by = "grp")+
theme_bw() +
theme(axis.text = element_blank(), axis.ticks = element_blank(),
panel.grid.minor = element_blank()) +
xlab("UMAP1") +
ylab("UMAP2")
origin
## visualize input data
DimPlot(seurat, reduction = "umap", cols=colOri, group.by = "origin")+
theme_bw() +
theme(axis.text = element_blank(), axis.ticks = element_blank(),
panel.grid.minor = element_blank()) +
xlab("UMAP1") +
ylab("UMAP2")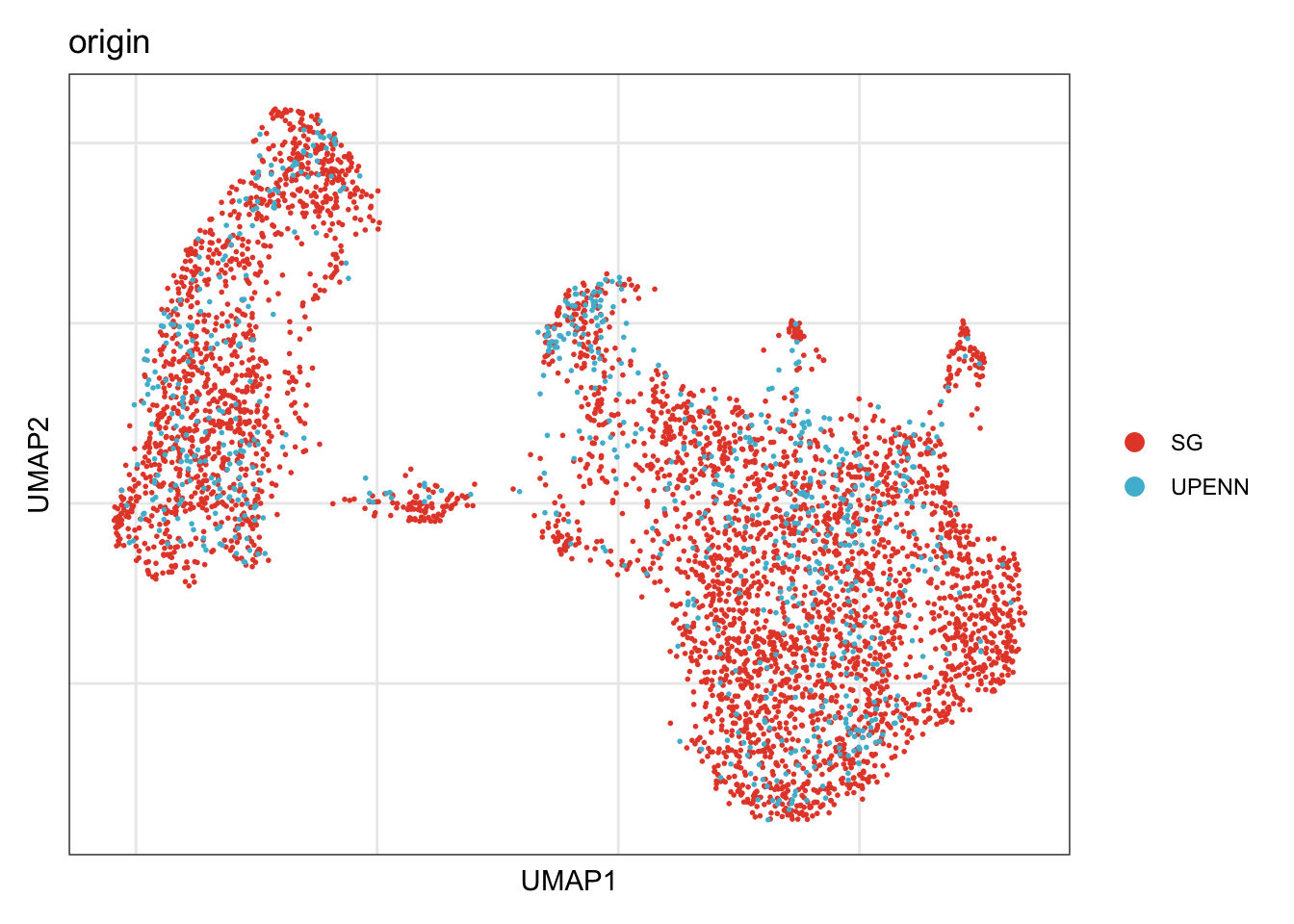
vis cluster marker
marker genes
seurat_markers_all <- FindAllMarkers(object = seurat, assay ="RNA",
only.pos = TRUE, min.pct = 0.25,
logfc.threshold = 0.25,
test.use = "wilcox")avg heatmap
cluster <- levels(seurat)
selGenesAll <- seurat_markers_all %>% group_by(cluster) %>%
top_n(-20, p_val_adj) %>%
top_n(20, avg_log2FC)
selGenesAll <- selGenesAll %>% mutate(geneIDval=gsub("^.*\\.", "", gene)) %>% filter(nchar(geneIDval)>1)
Idents(seurat) <- seurat$intCluster
pOut <- avgHeatmap(seurat = seurat, selGenes = selGenesAll,
colVecIdent = colPal,
ordVec=levels(seurat),
gapVecR=NULL, gapVecC=NULL,cc=FALSE,
cr=T, condCol=F)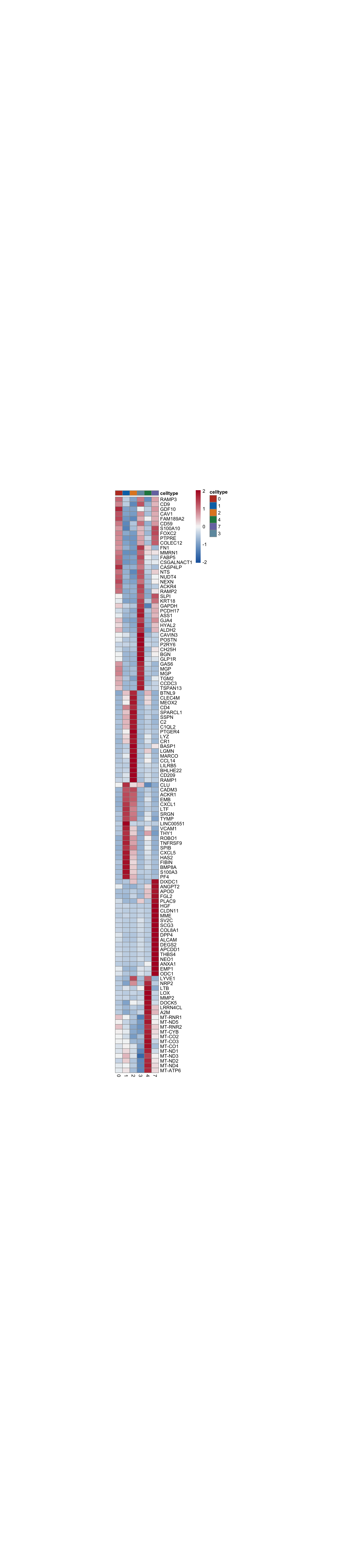
write table
write.table(seurat_markers_all,
file=paste0(basedir,
"/data/AllPatWithoutCM_LEConly_intOrig_markerGenes.txt"),
row.names = FALSE, col.names = TRUE, quote = FALSE, sep = "\t")asign labels
seurat$label <- "SCScLEC"
seurat$label[which(seurat$intCluster %in% c("1"))] <- "SCSfLEC"
seurat$label[which(seurat$intCluster %in% c("2"))] <- "MedSinusLEC"
seurat$label[which(seurat$intCluster %in% c("3"))] <- "MedCapsLEC"
seurat$label[which(seurat$intCluster %in% c("4"))] <- "ParacortLEC"
seurat$label[which(seurat$intCluster %in% c("7"))] <- "ValveLEC"visualize label
Idents(seurat) <- seurat$label
DimPlot(seurat, reduction = "umap", cols=colLab)+
theme_bw() +
theme(axis.text = element_blank(), axis.ticks = element_blank(),
panel.grid.minor = element_blank()) +
xlab("UMAP1") +
ylab("UMAP2")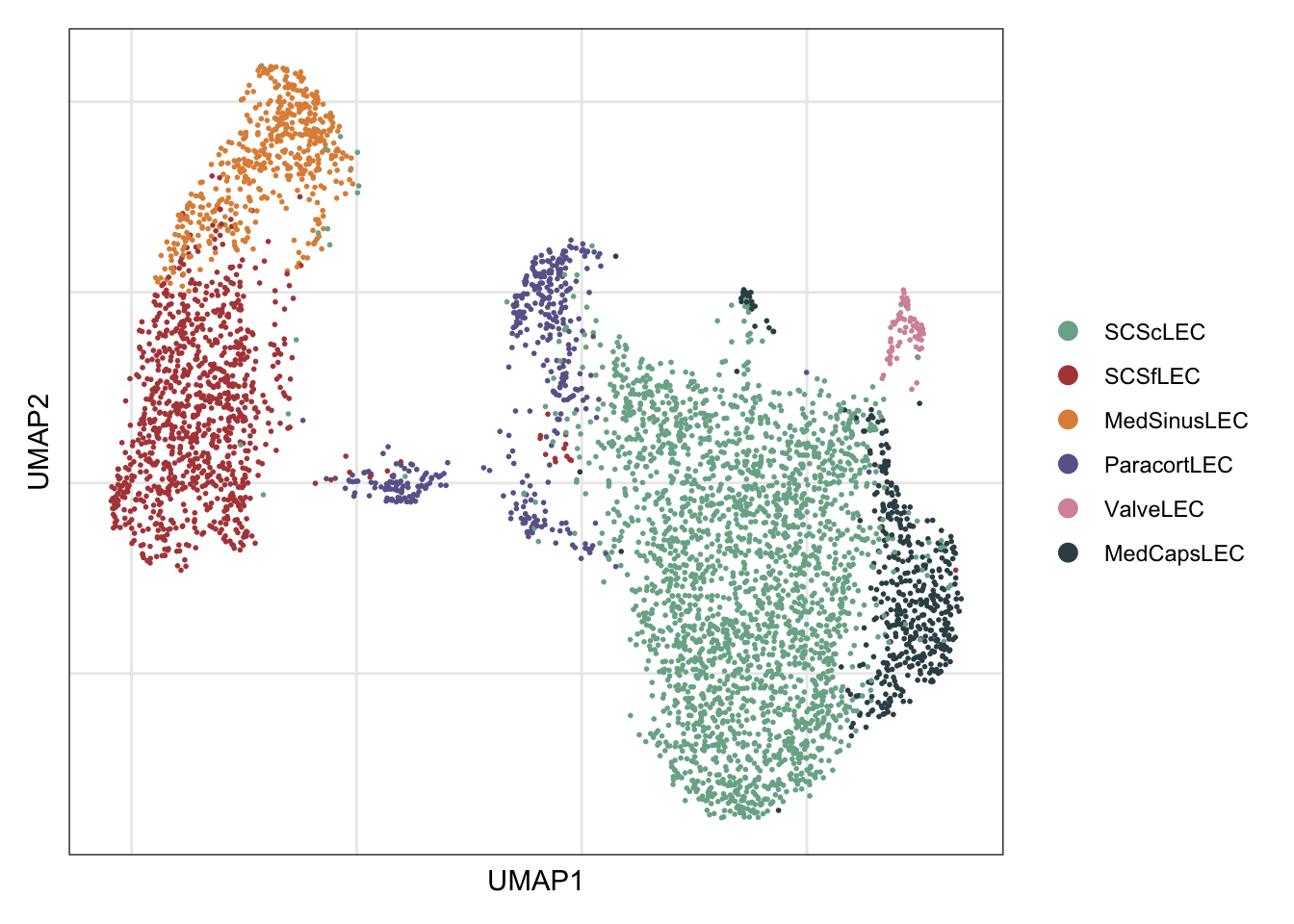
DimPlot(seurat, reduction = "umap", cols=colLab, pt.size=0.5)+
theme_void()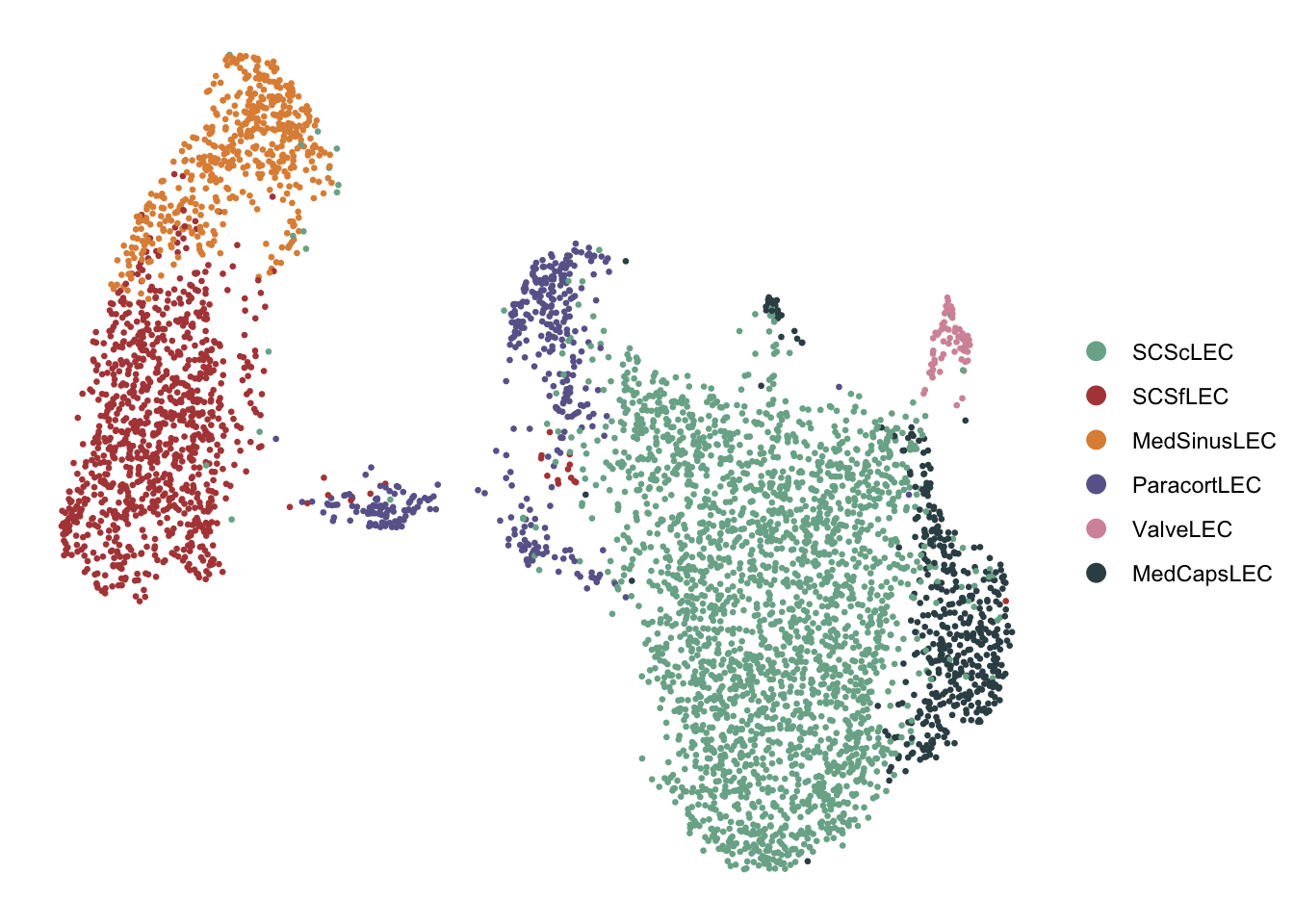
vis selected LEC marker
genes <- data.frame(gene=rownames(seurat)) %>%
mutate(geneID=gsub("^.*\\.", "", gene))
selGenesAll <- read_tsv(file = paste0(basedir,
"/data/overallLECMarker.txt")) %>%
left_join(., genes, by = "geneID")
seurat$label <- factor(seurat$label, levels = names(colLab))
Idents(seurat) <- seurat$label
pOut <- avgHeatmap(seurat = seurat, selGenes = selGenesAll,
colVecIdent = colLab,
ordVec=levels(seurat),
gapVecR=NULL, gapVecC=NULL,cc=F,
cr=F, condCol=F)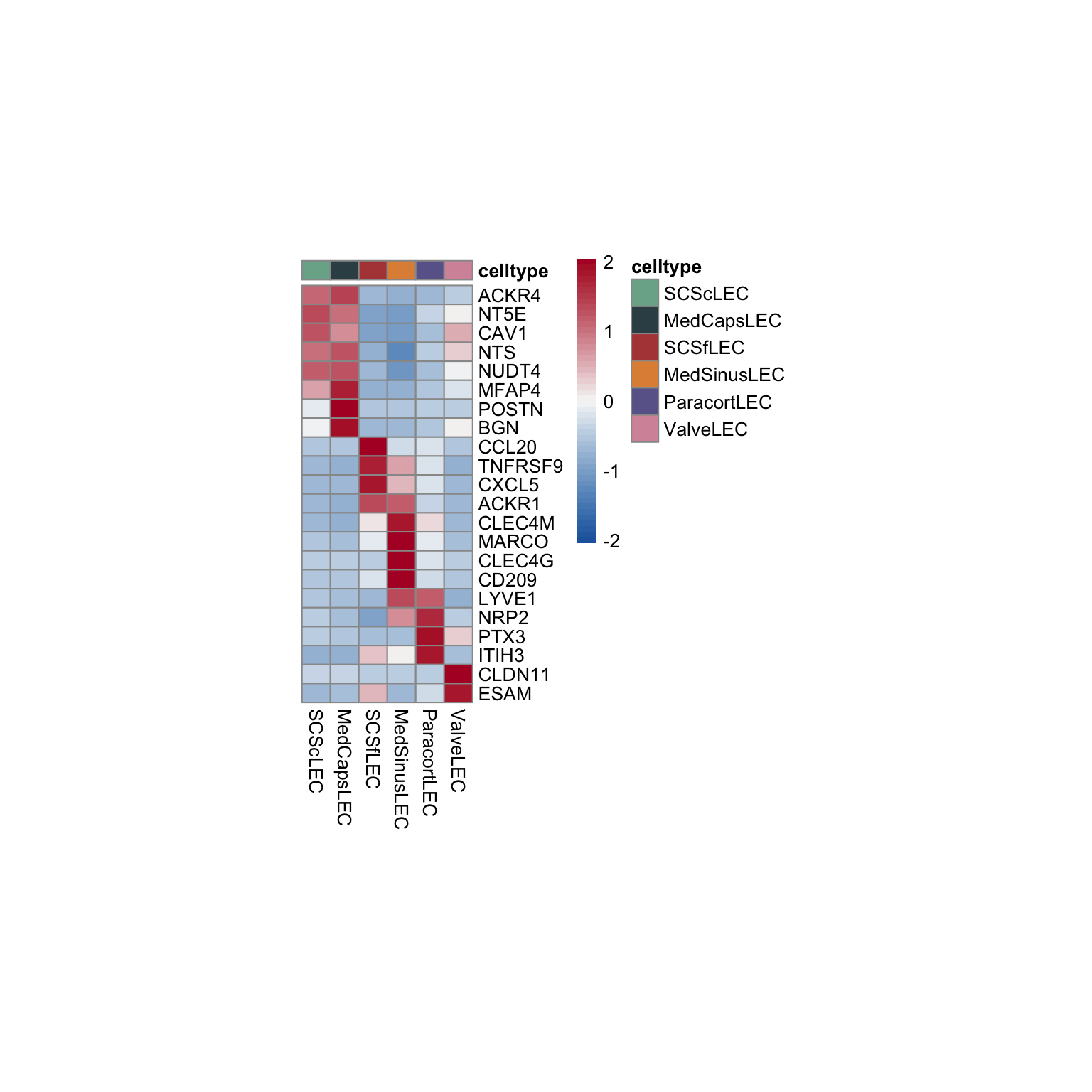
Dotplot
DotPlot(seurat, assay="RNA", features = rev(selGenesAll$gene), scale =T,
cluster.idents = F) +
scale_color_viridis_c() +
coord_flip() +
theme(axis.text.x = element_text(angle = 90, hjust = 1)) +
scale_x_discrete(breaks=rev(selGenesAll$gene), labels=rev(selGenesAll$geneID)) +
xlab("") + ylab("")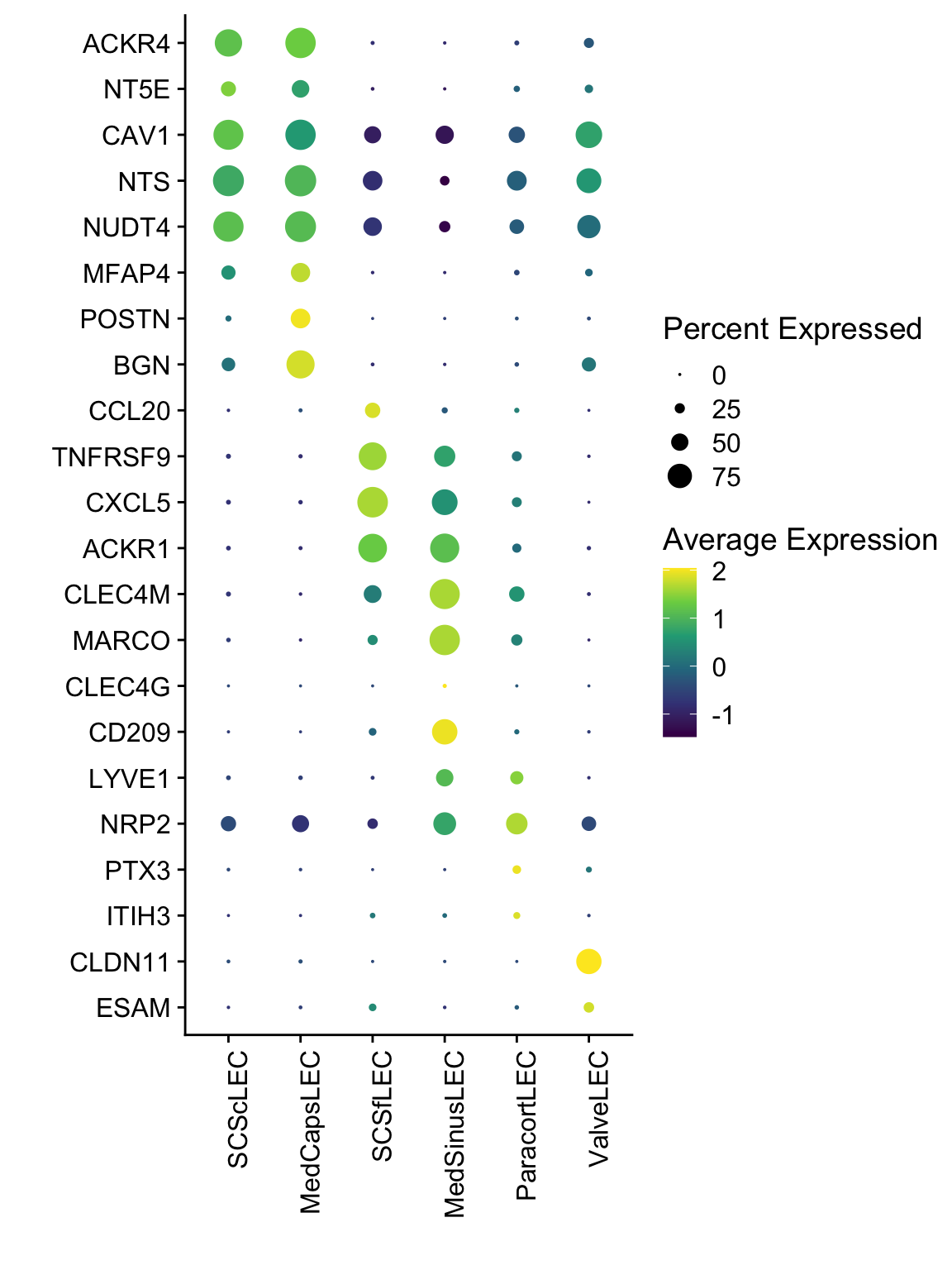
DotPlot(seurat, assay="RNA", features = rev(selGenesAll$gene), scale =F,
cluster.idents = F) +
scale_color_viridis_c() +
coord_flip() +
theme(axis.text.x = element_text(angle = 90, hjust = 1)) +
scale_x_discrete(breaks=rev(selGenesAll$gene), labels=rev(selGenesAll$geneID)) +
xlab("") + ylab("")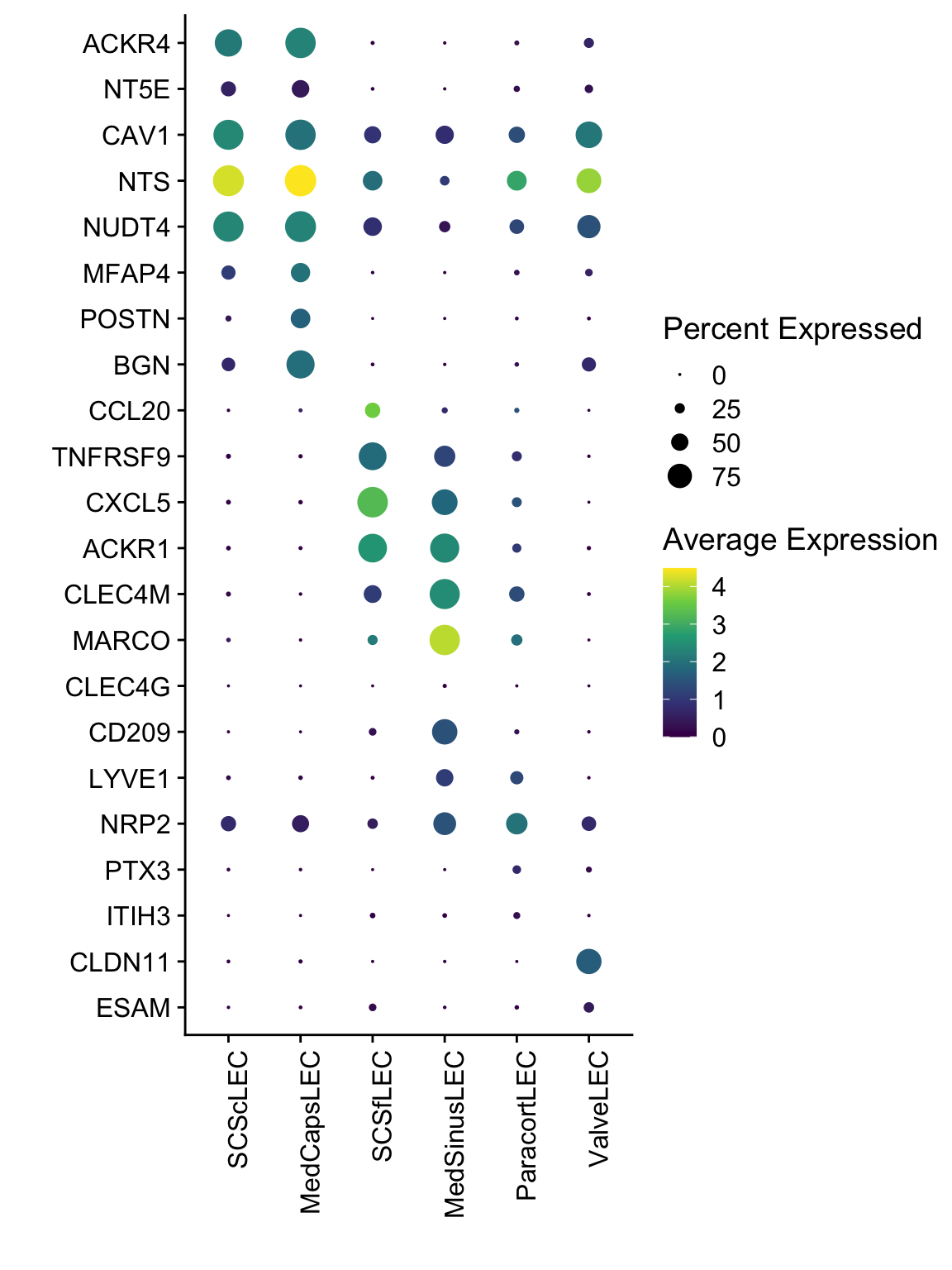
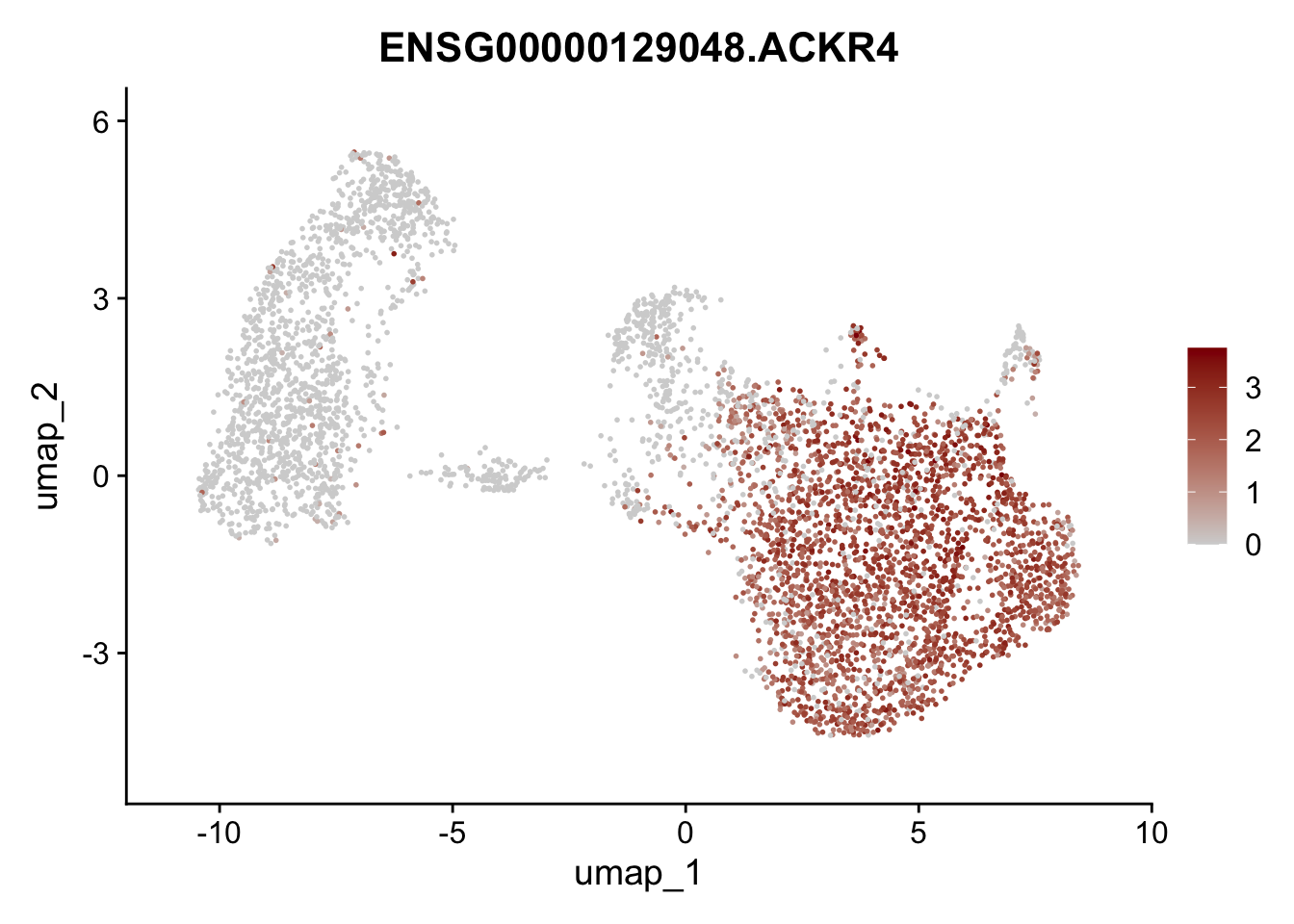
Featureplot
pList <- sapply(selGenesAll$gene, function(x){
p <- FeaturePlot(seurat, reduction = "umap",
features = x,
cols=c("lightgrey", "darkred"),
order = F)+
theme(legend.position="right")
plot(p)
})
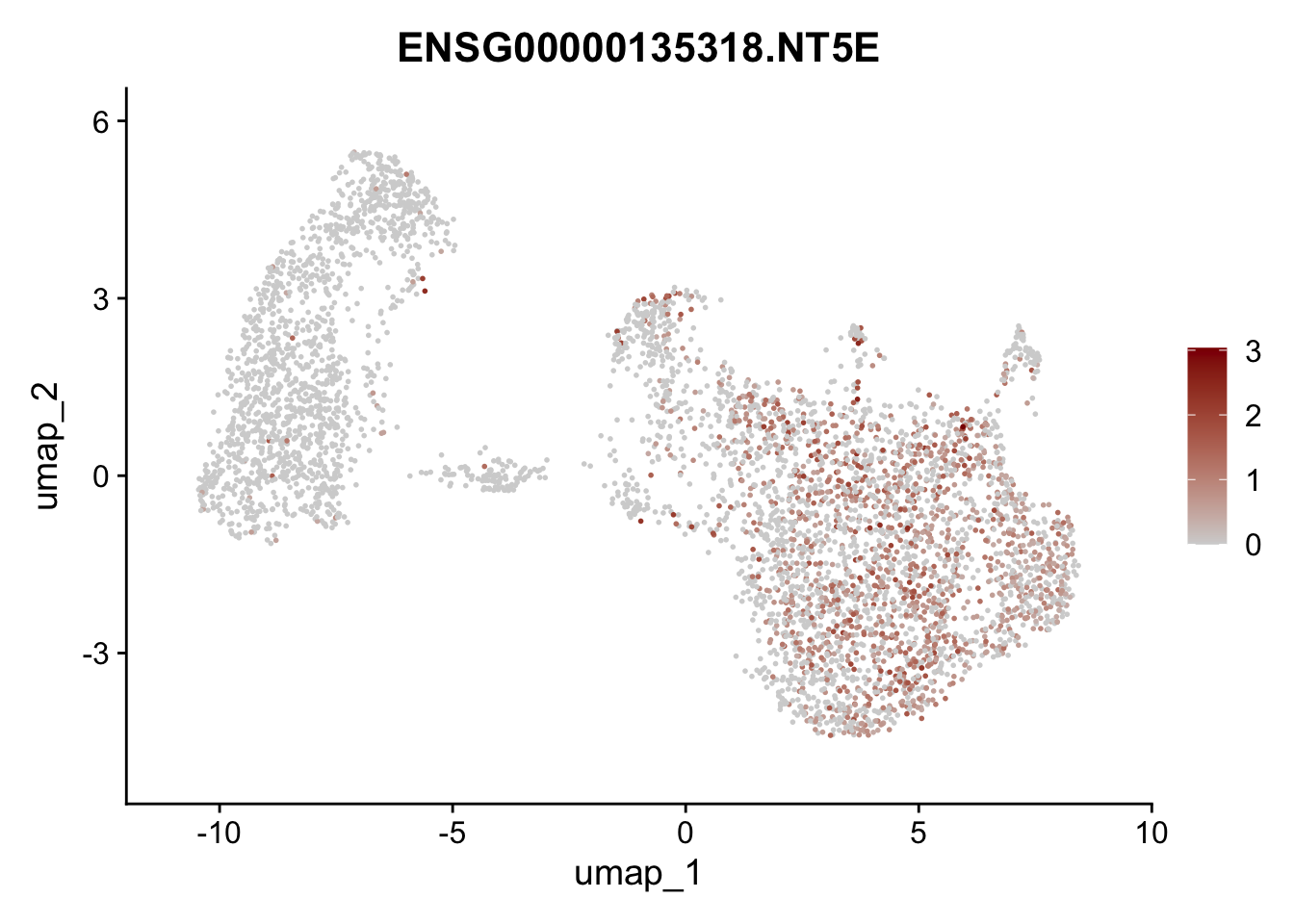
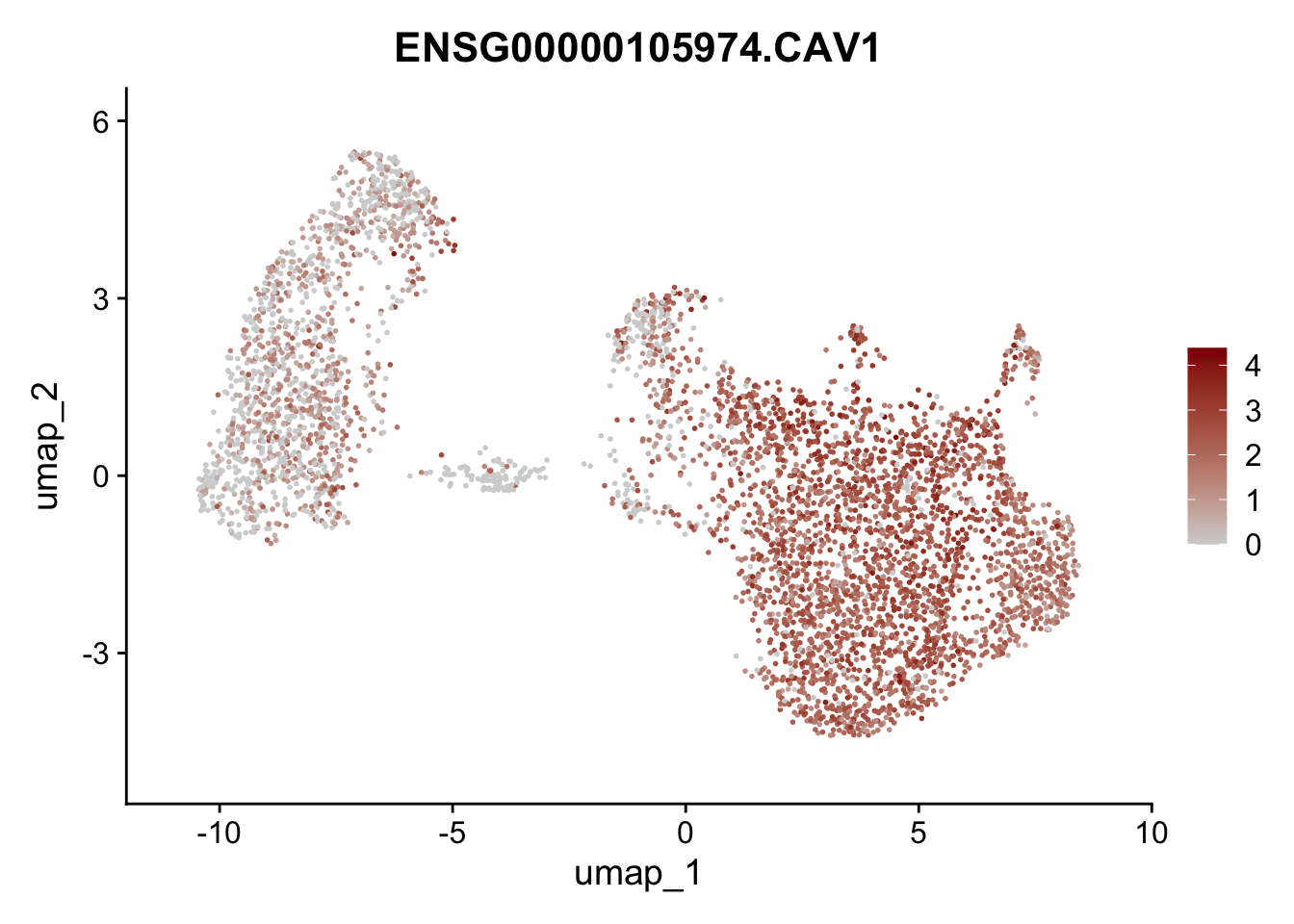
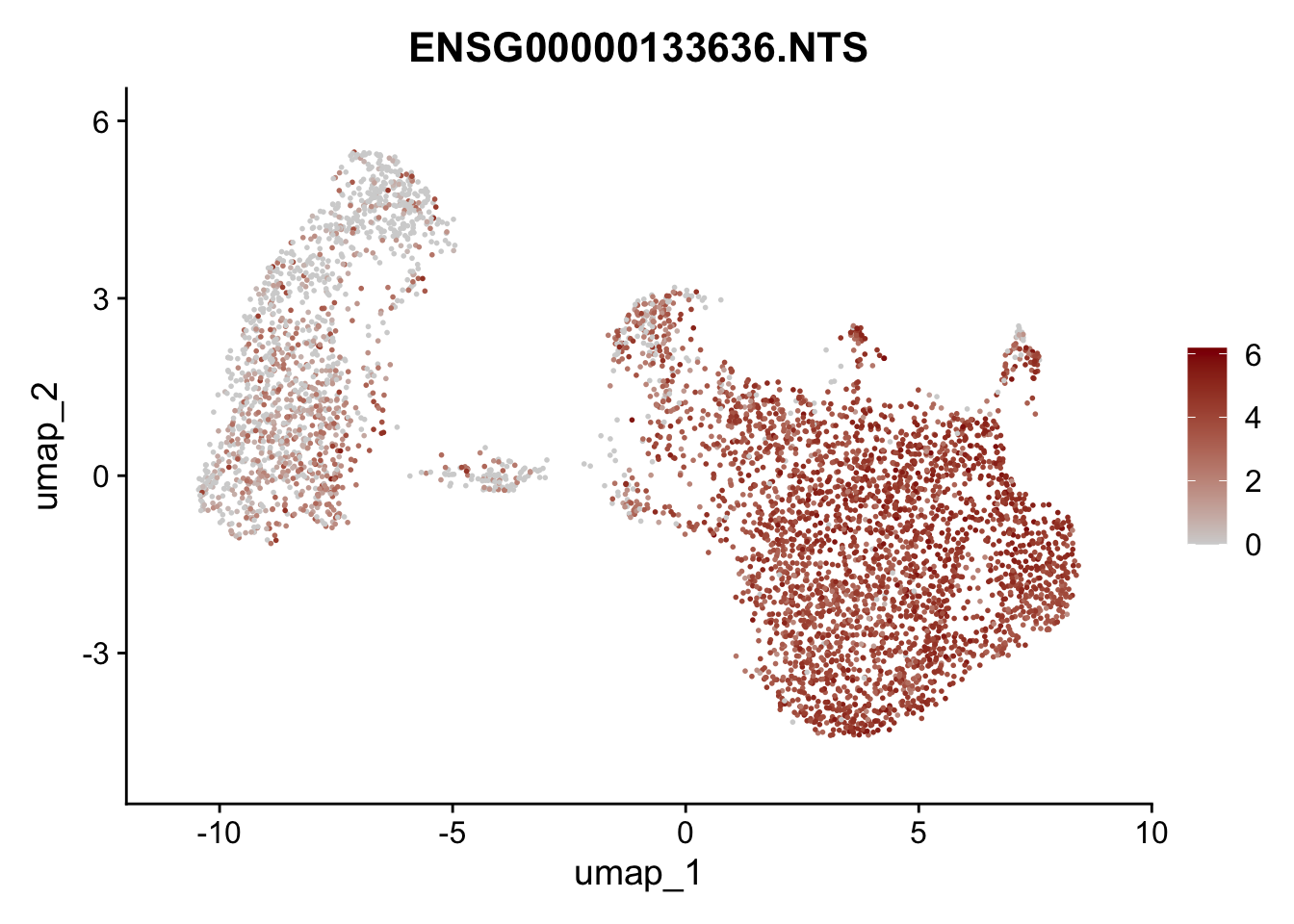
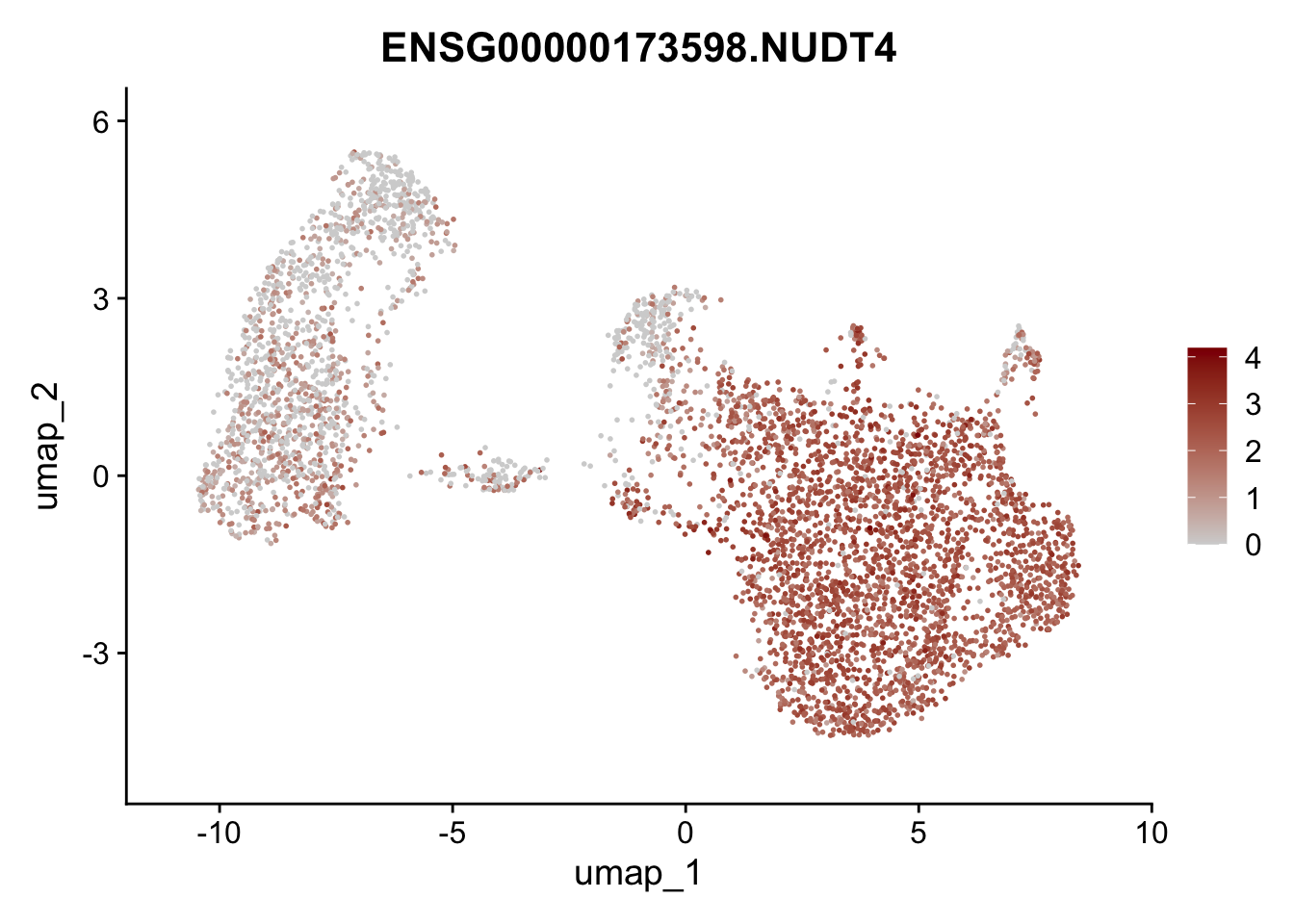
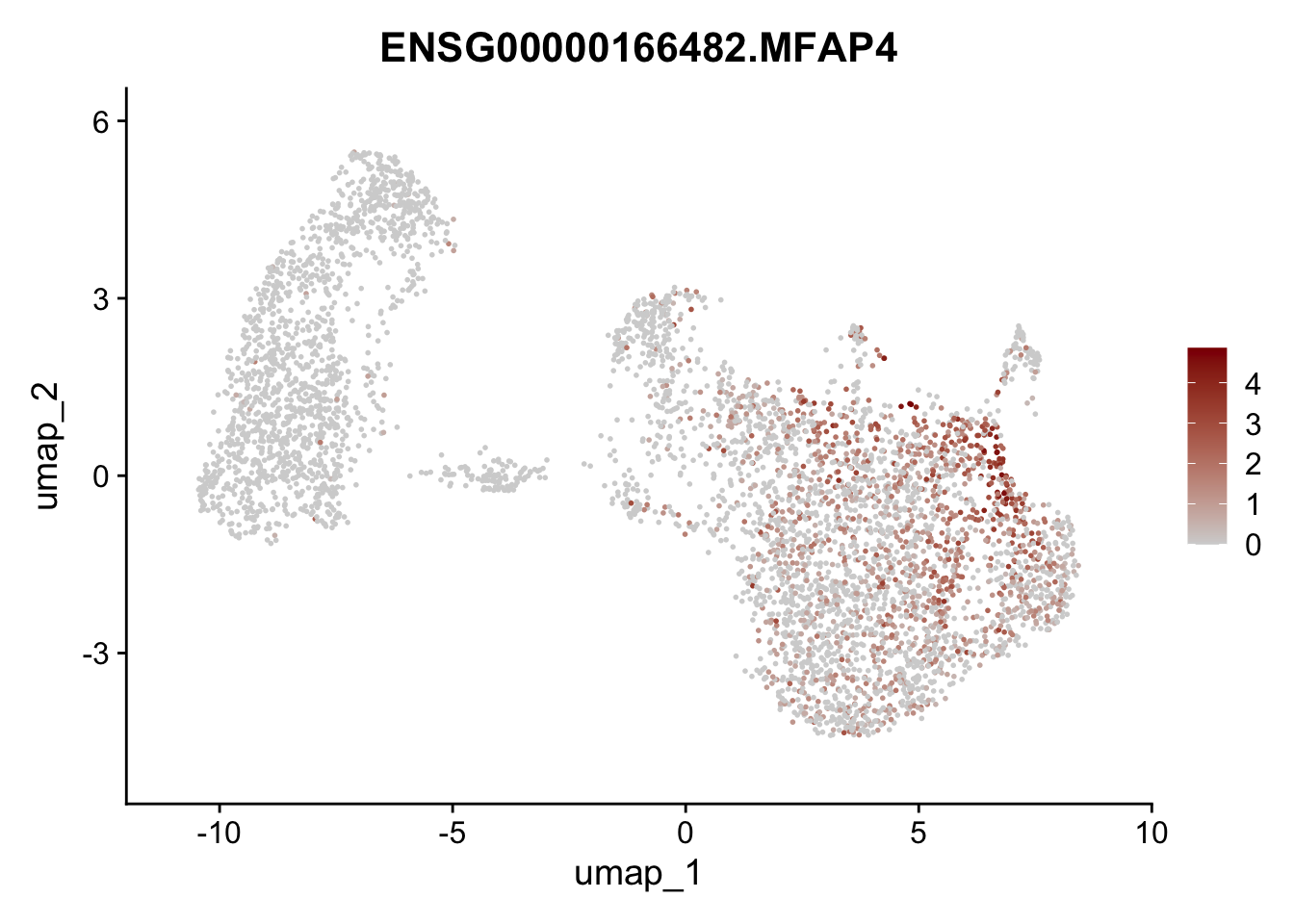
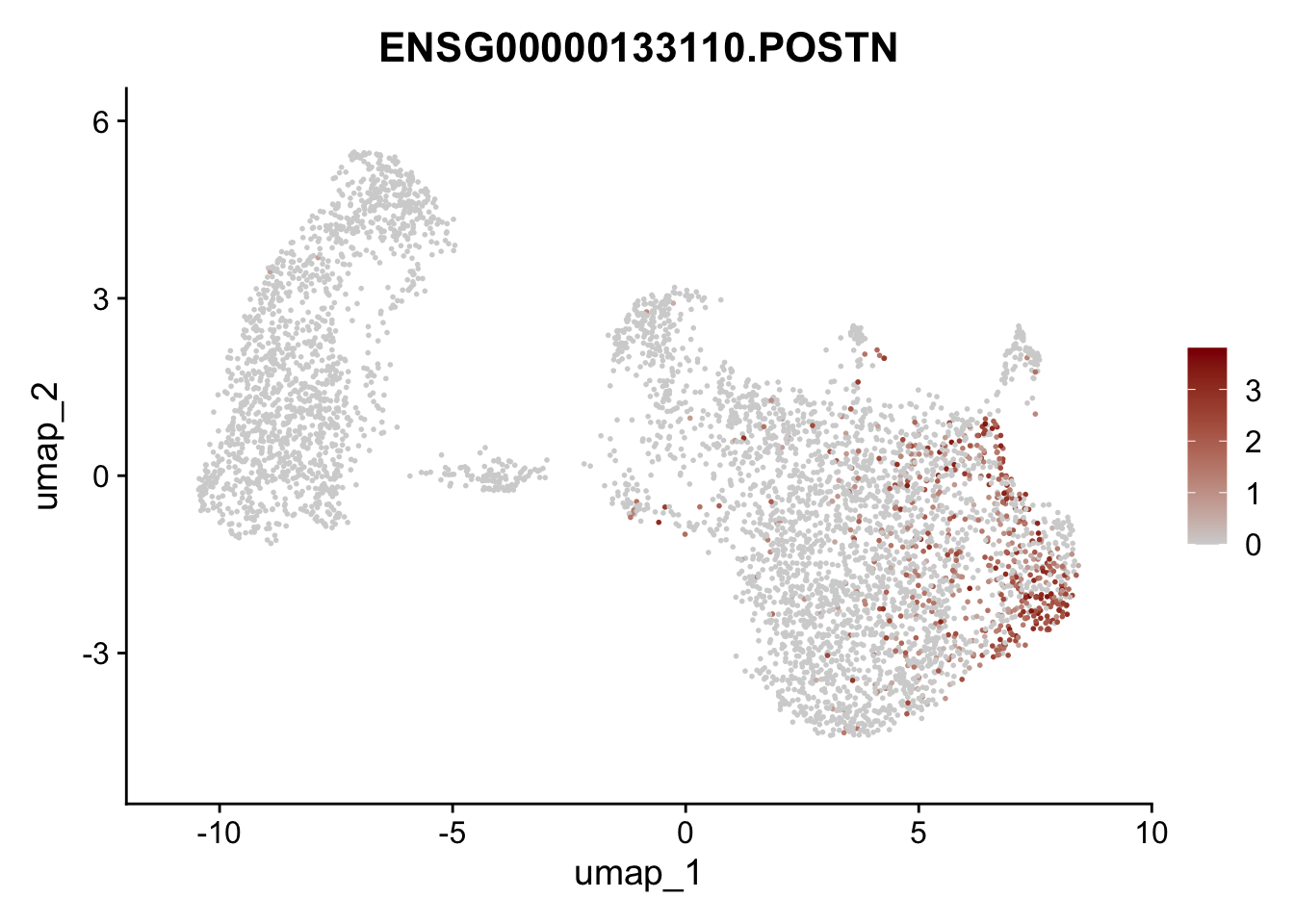
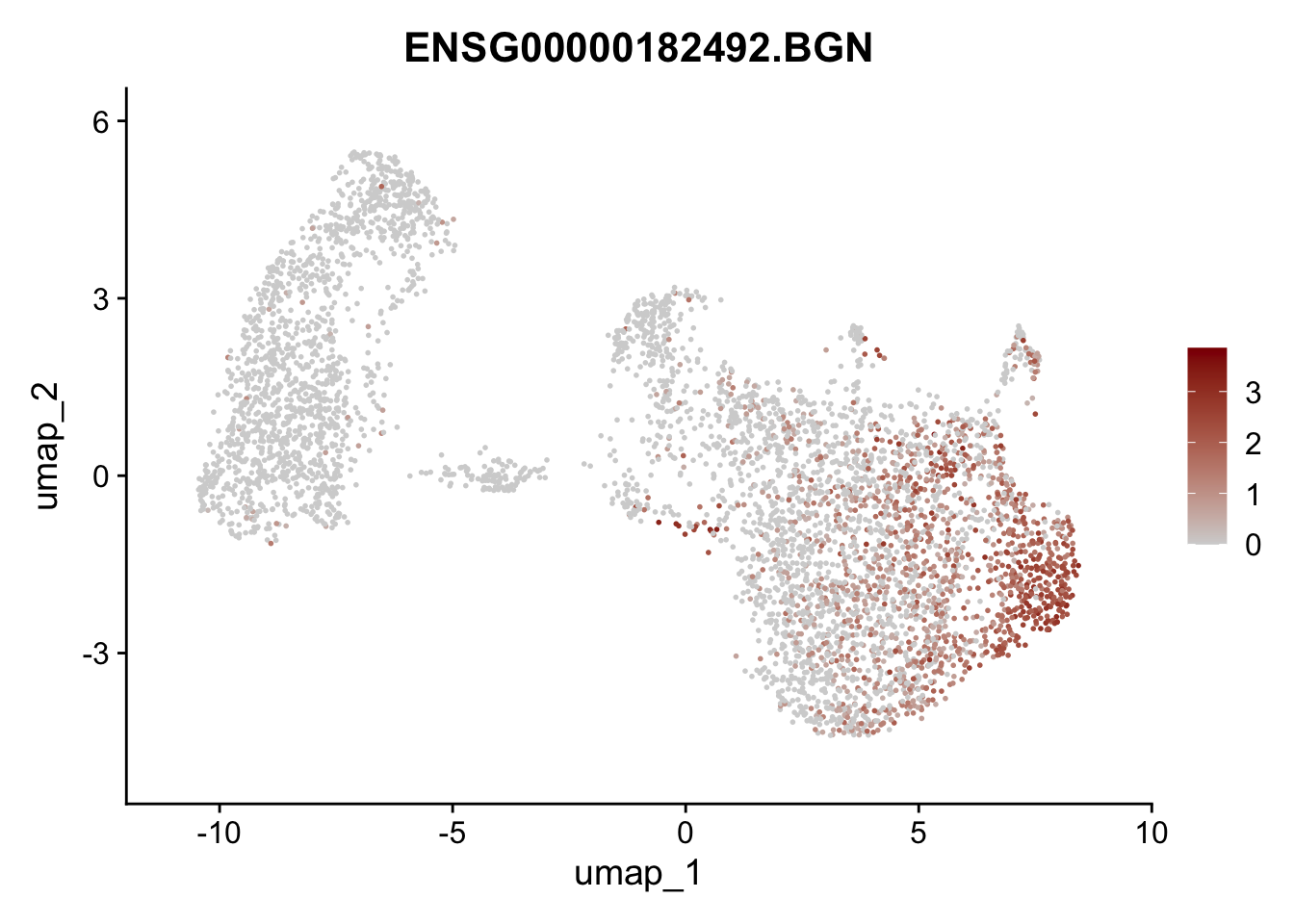
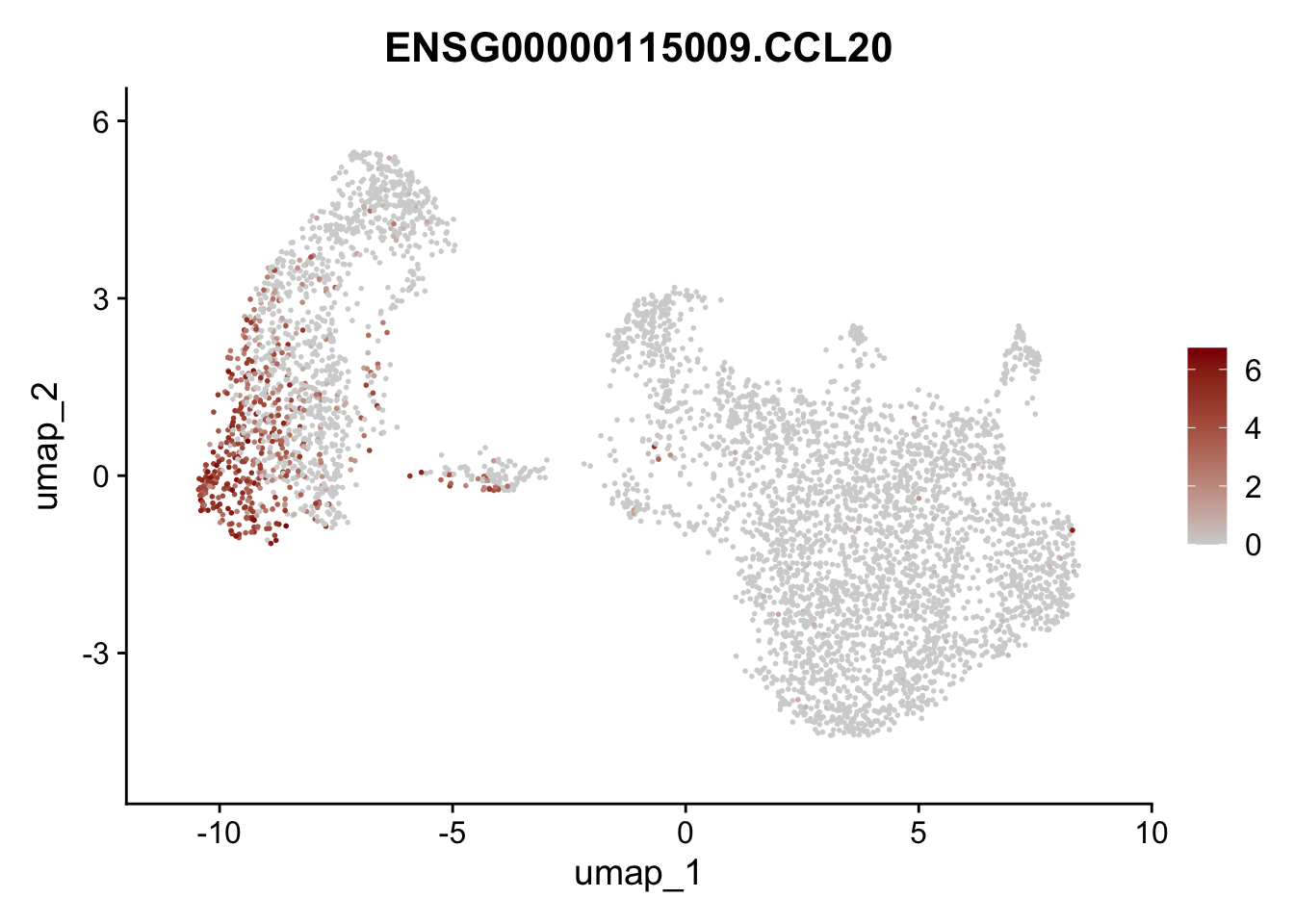
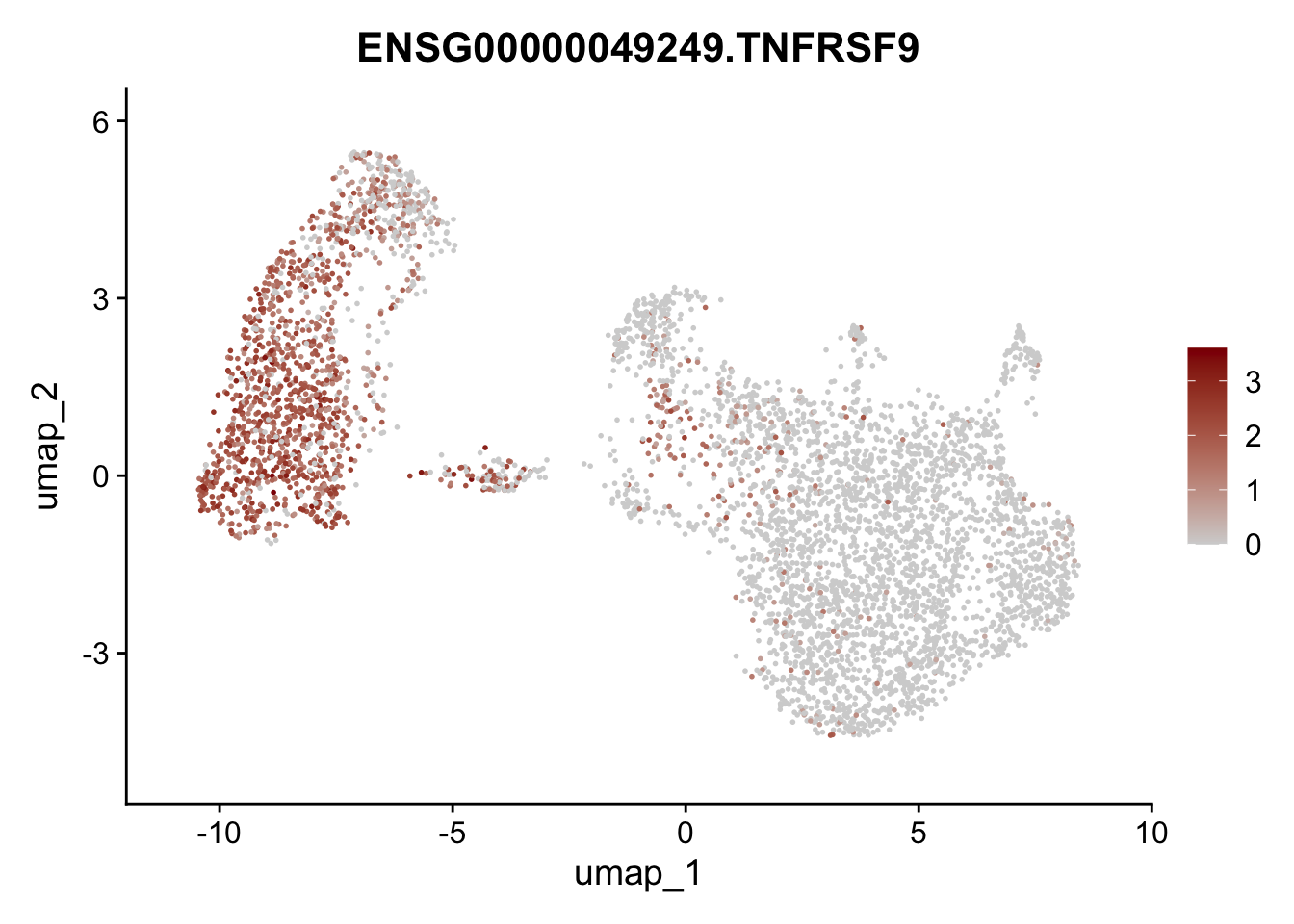
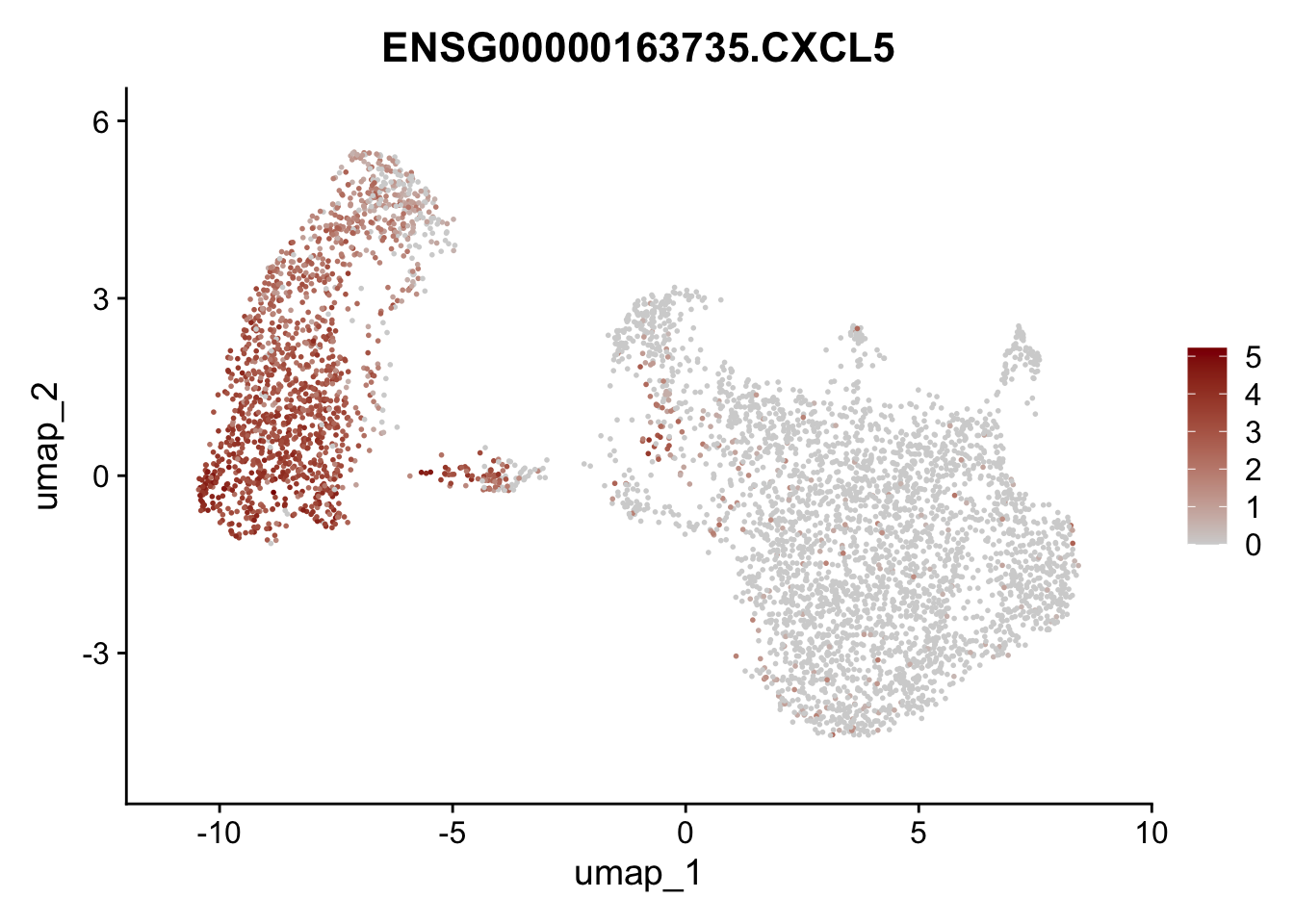
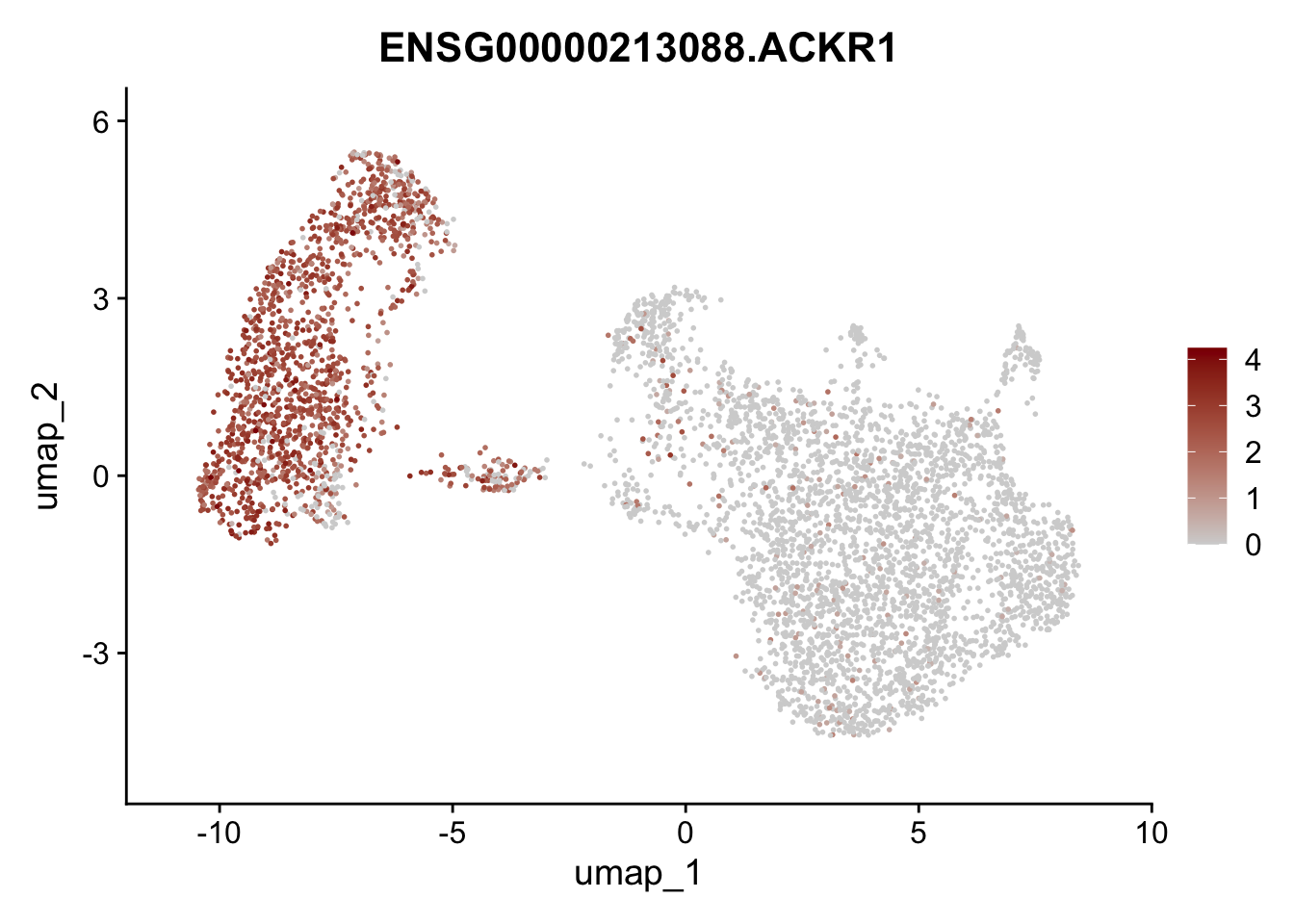
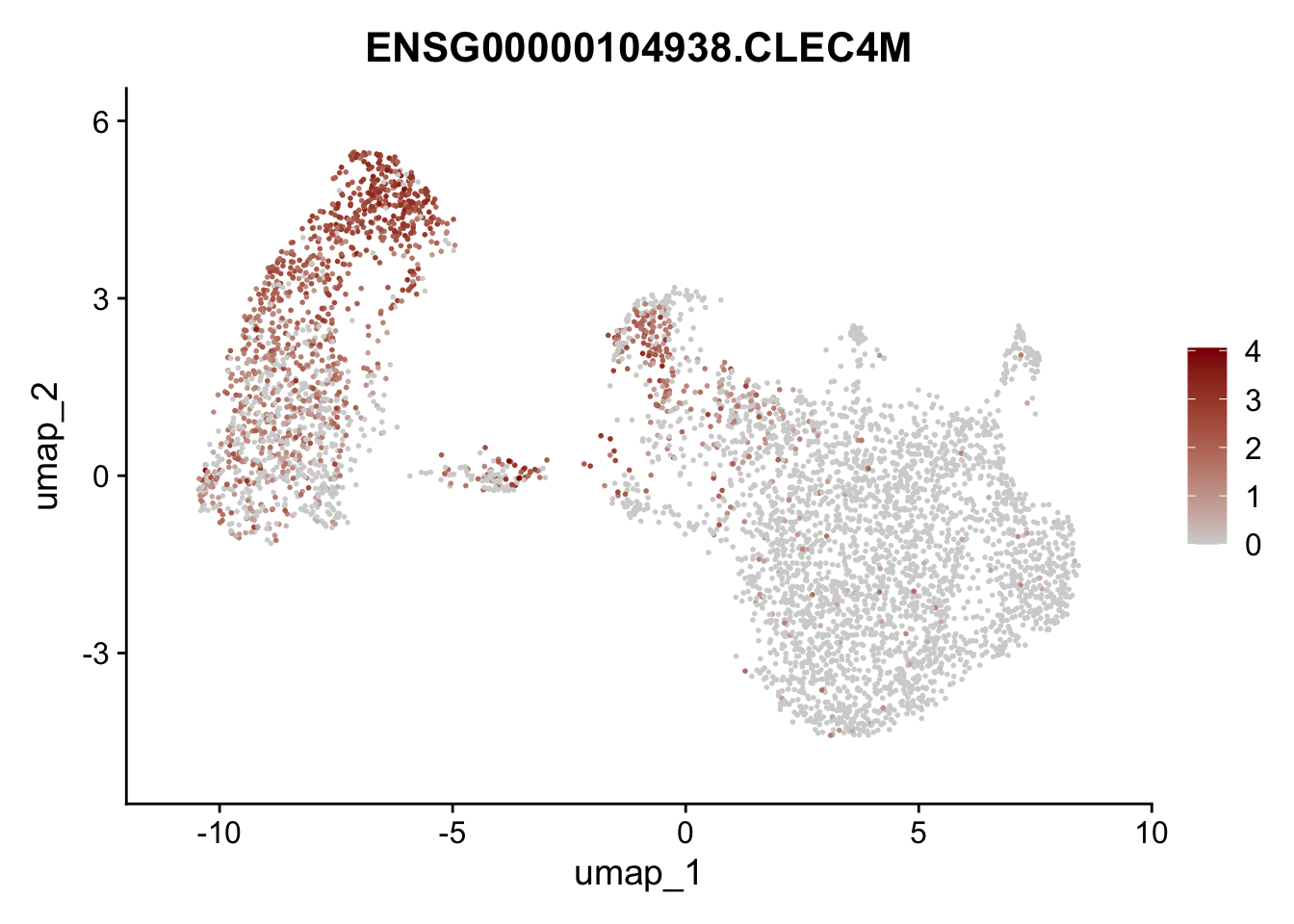
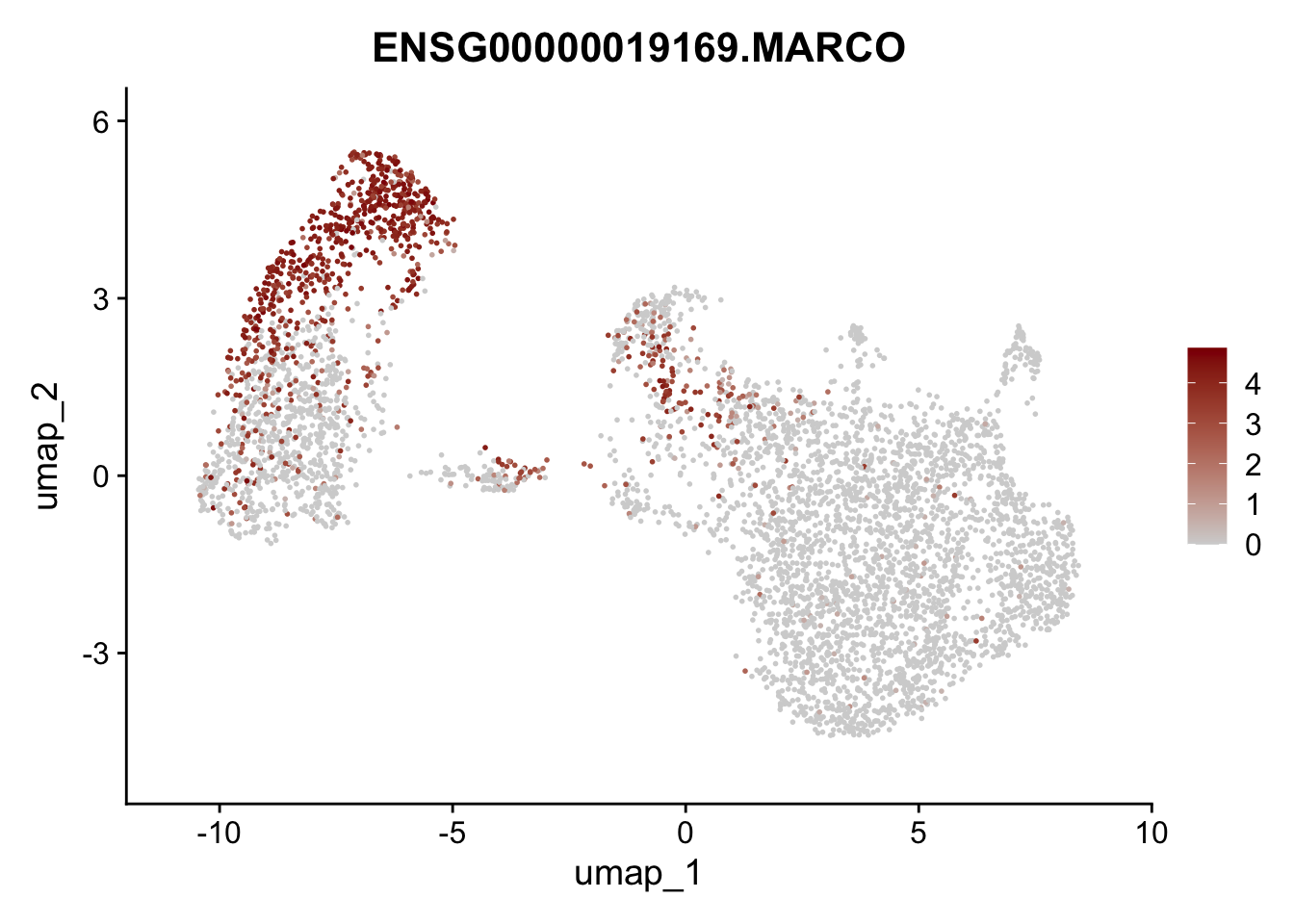
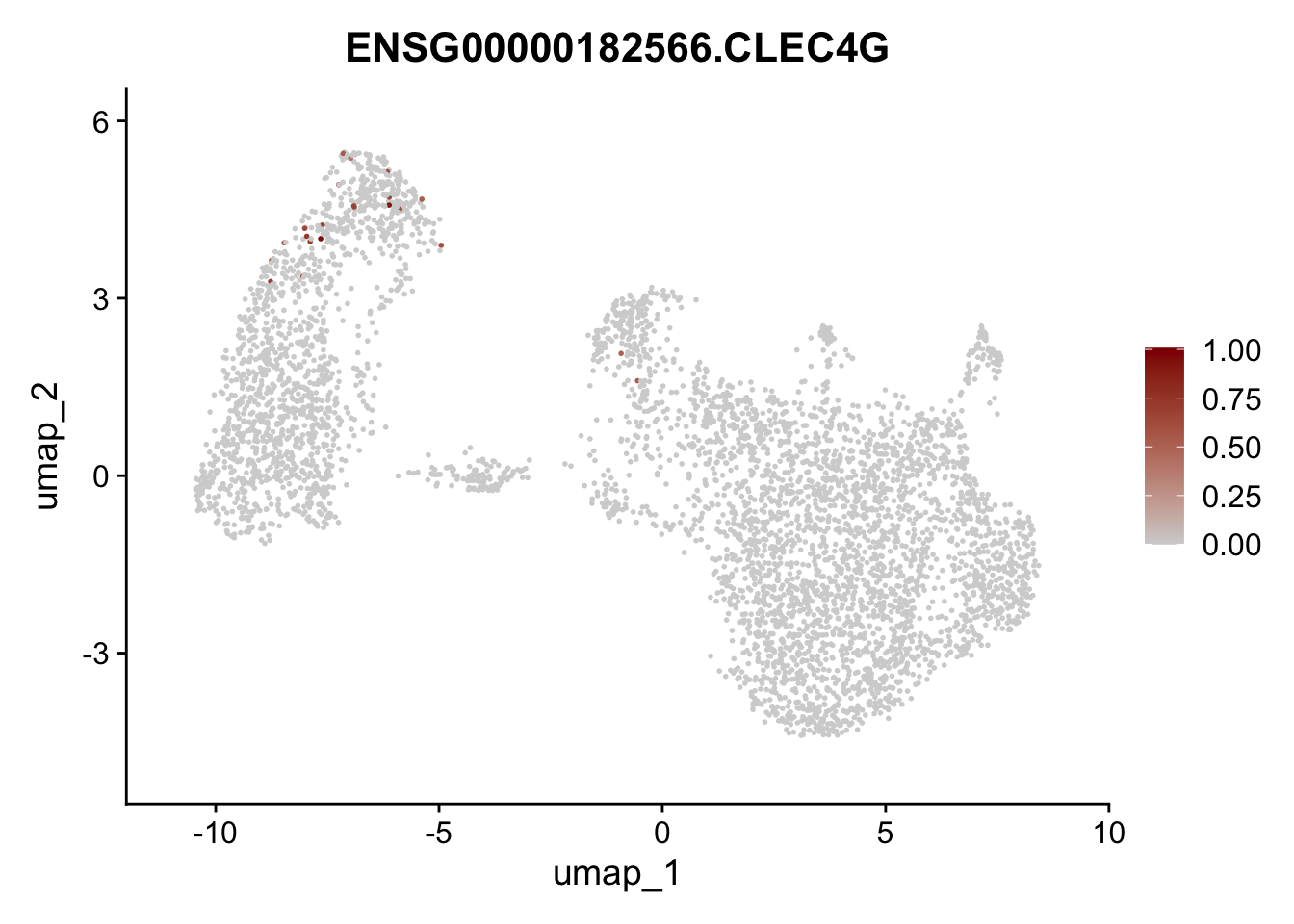
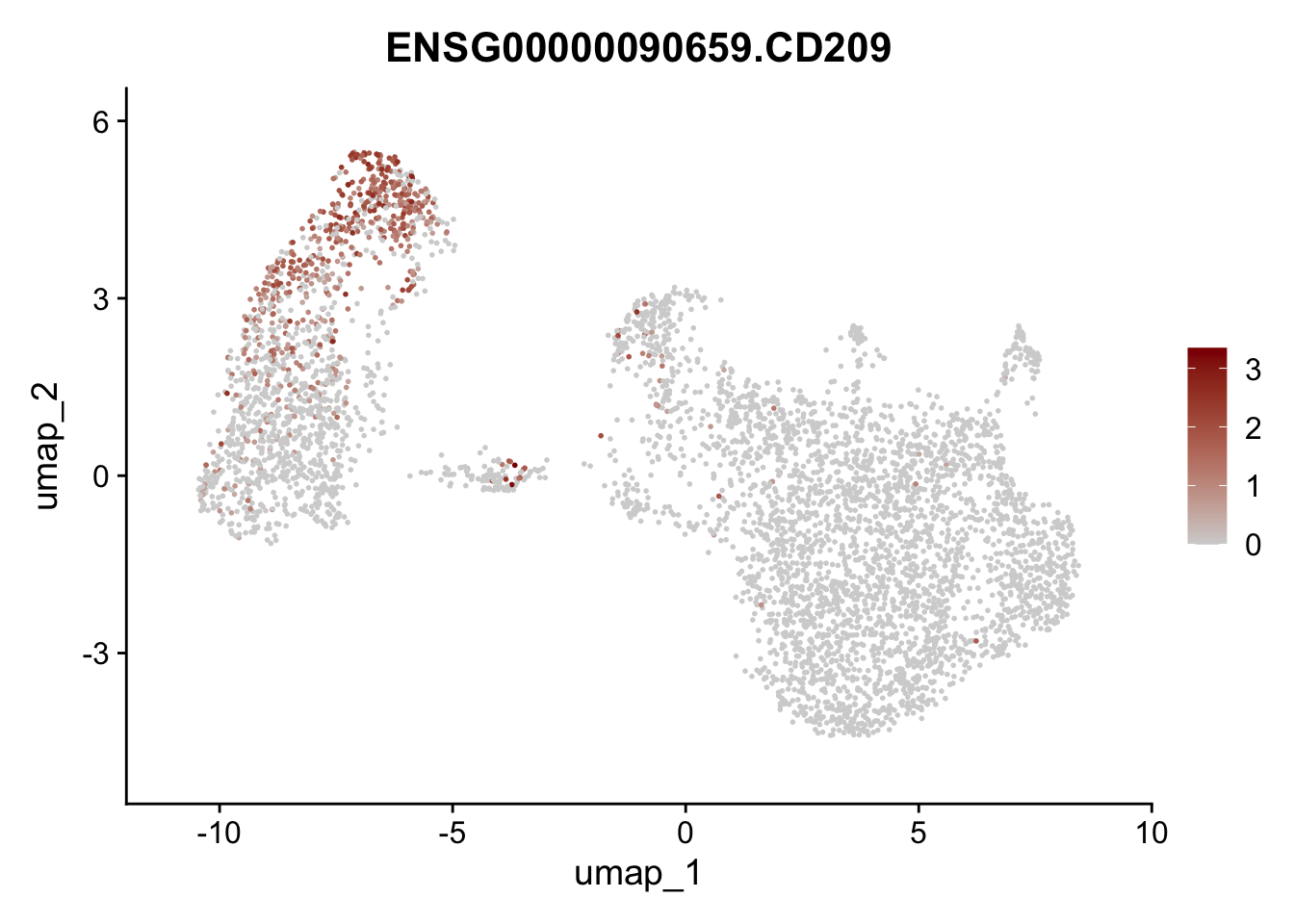
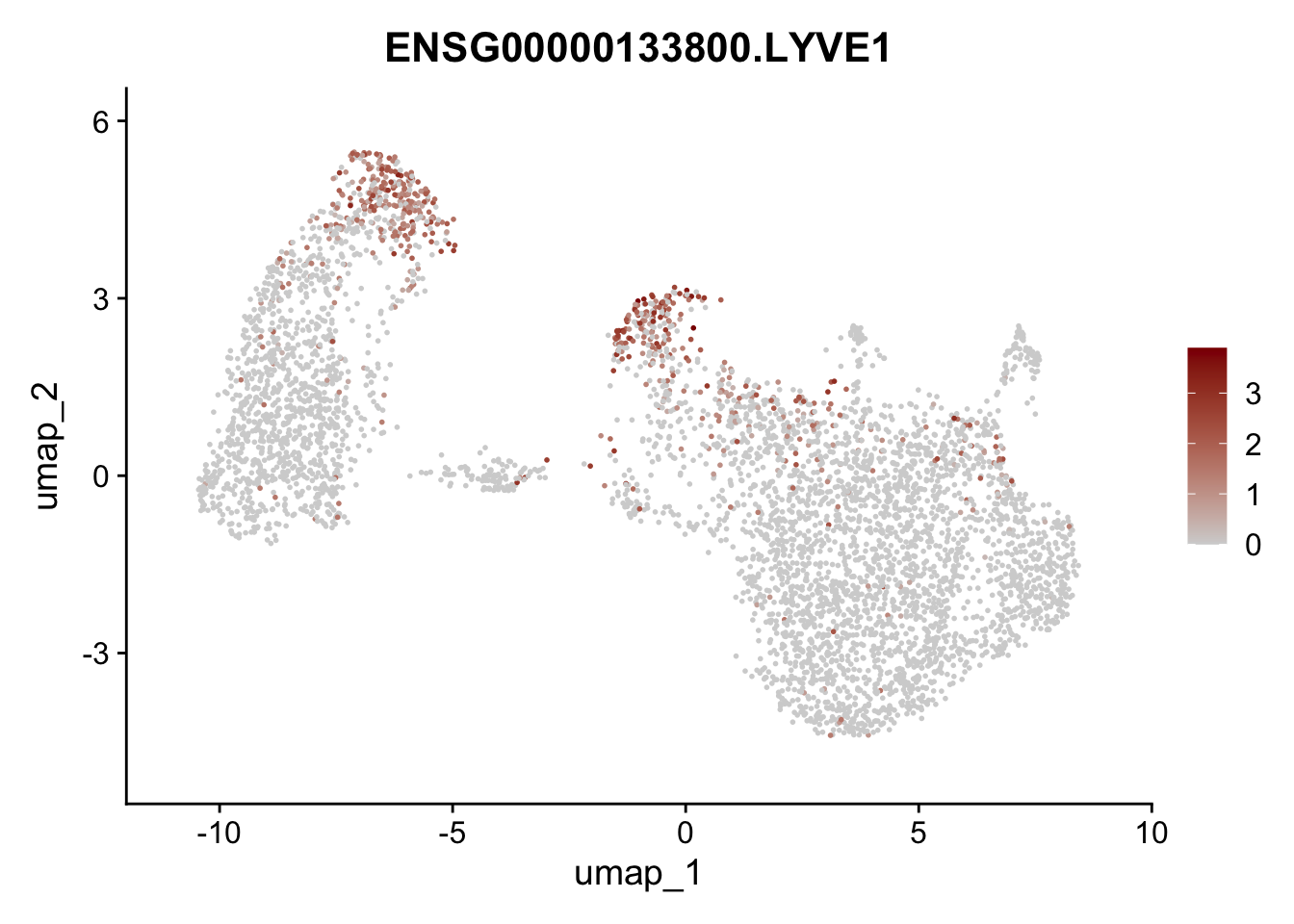
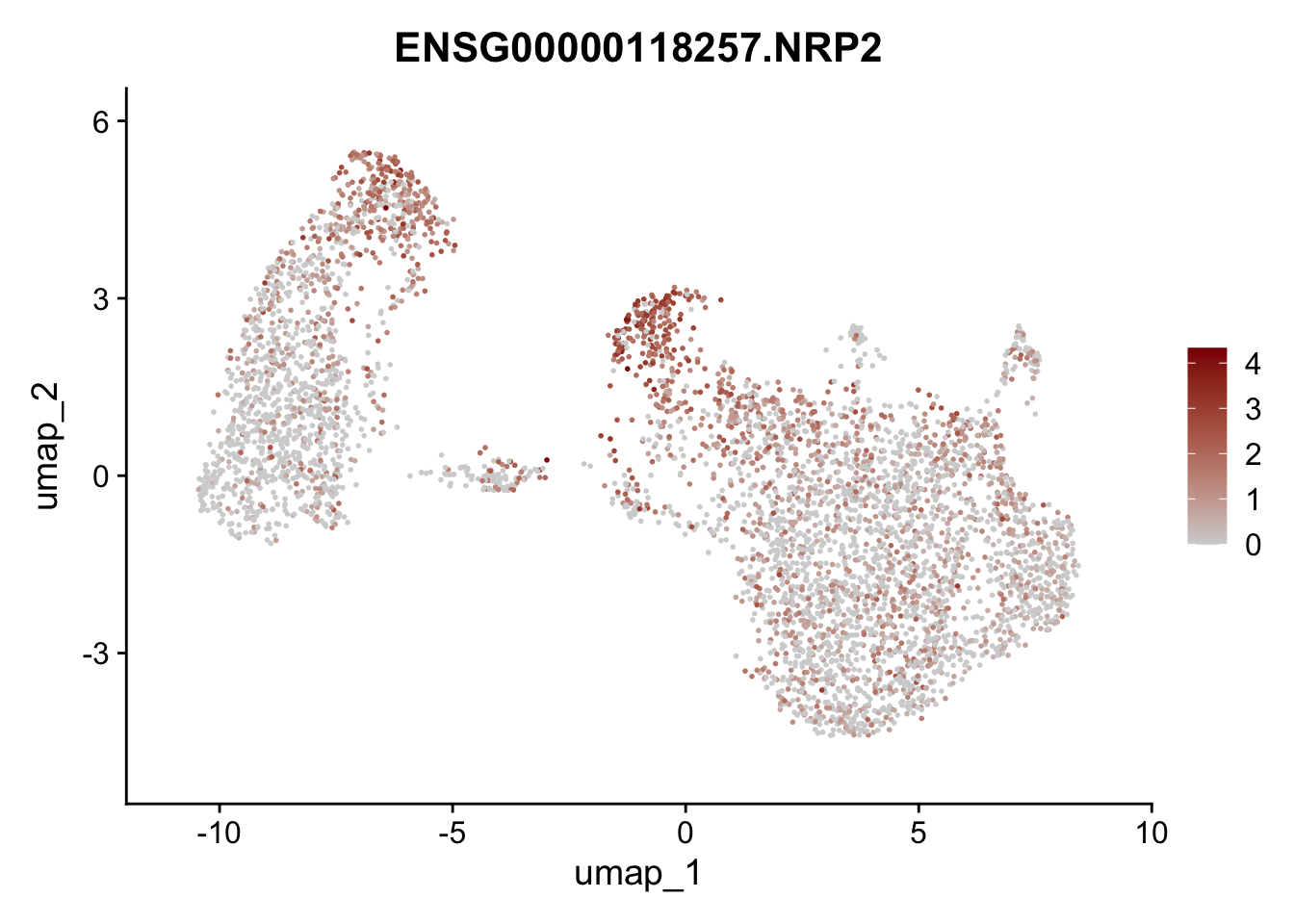
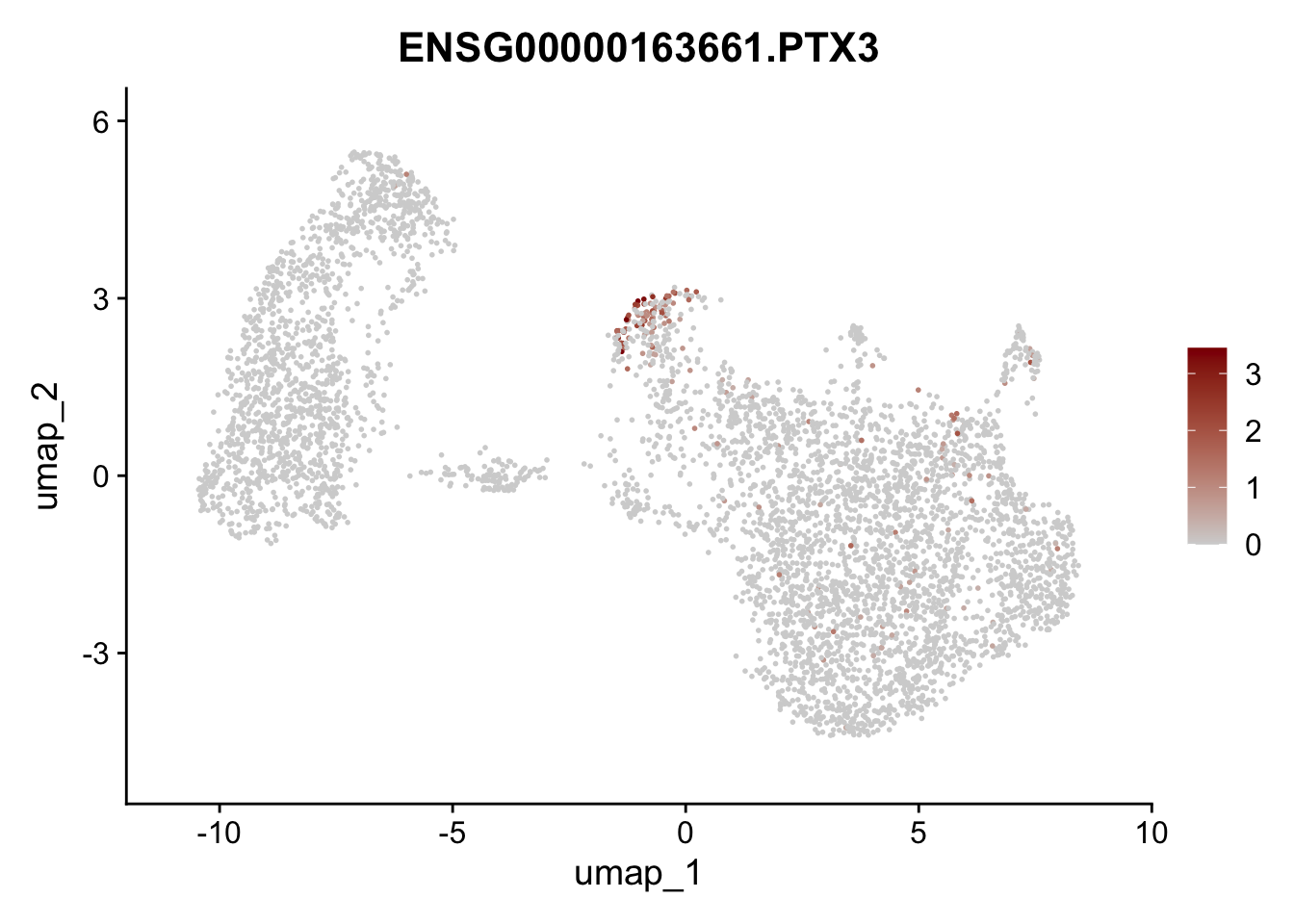
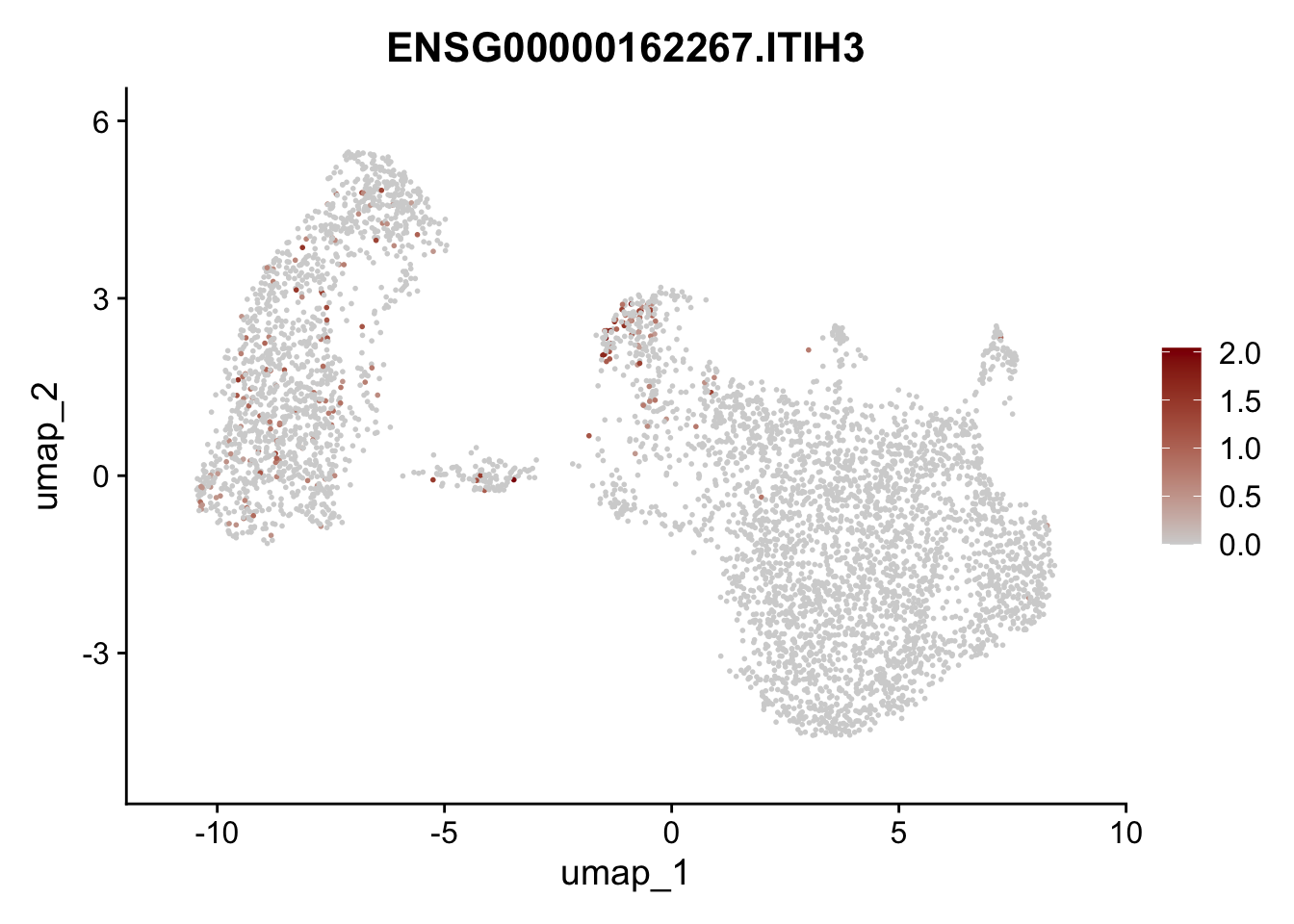
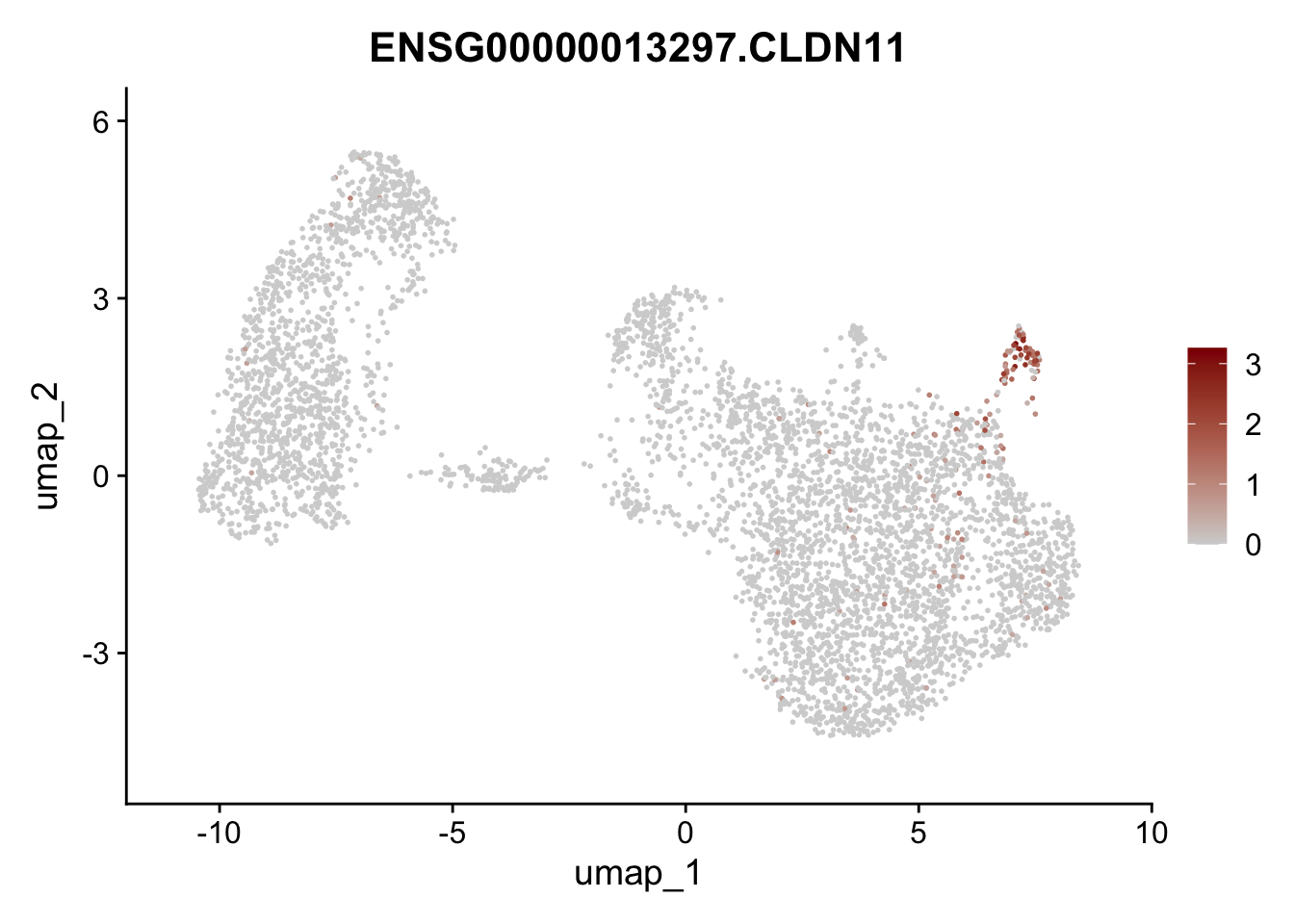
save labeled seurat object
saveRDS(seurat, paste0(basedir,
"/data/AllPatWithoutCM_LEConly_intOrig_label",
"_seurat.rds"))session info
sessionInfo()R version 4.3.0 (2023-04-21)
Platform: x86_64-apple-darwin20 (64-bit)
Running under: macOS Ventura 13.4.1
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.3-x86_64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.3-x86_64/Resources/lib/libRlapack.dylib; LAPACK version 3.11.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: Europe/Berlin
tzcode source: internal
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods base
other attached packages:
[1] scater_1.28.0 scuttle_1.10.3 SingleCellExperiment_1.22.0
[4] SummarizedExperiment_1.30.2 Biobase_2.60.0 GenomicRanges_1.52.1
[7] GenomeInfoDb_1.36.4 IRanges_2.36.0 S4Vectors_0.40.1
[10] BiocGenerics_0.48.0 MatrixGenerics_1.12.3 matrixStats_1.2.0
[13] pheatmap_1.0.12 ggsci_3.0.1 here_1.0.1
[16] runSeurat3_0.1.0 ggpubr_0.6.0 lubridate_1.9.3
[19] forcats_1.0.0 stringr_1.5.1 readr_2.1.5
[22] tidyr_1.3.1 tibble_3.2.1 tidyverse_2.0.0
[25] Seurat_5.0.2 SeuratObject_5.0.1 sp_2.1-3
[28] purrr_1.0.2 cowplot_1.1.3 ggplot2_3.5.0
[31] reshape2_1.4.4 dplyr_1.1.4
loaded via a namespace (and not attached):
[1] RcppAnnoy_0.0.22 splines_4.3.0 later_1.3.2
[4] bitops_1.0-7 polyclip_1.10-6 fastDummies_1.7.3
[7] lifecycle_1.0.4 rstatix_0.7.2 rprojroot_2.0.4
[10] vroom_1.6.5 globals_0.16.2 lattice_0.22-5
[13] MASS_7.3-60.0.1 backports_1.4.1 magrittr_2.0.3
[16] limma_3.56.2 plotly_4.10.4 rmarkdown_2.26
[19] yaml_2.3.8 httpuv_1.6.14 sctransform_0.4.1
[22] spam_2.10-0 spatstat.sparse_3.0-3 reticulate_1.35.0
[25] pbapply_1.7-2 RColorBrewer_1.1-3 abind_1.4-5
[28] zlibbioc_1.46.0 Rtsne_0.17 presto_1.0.0
[31] RCurl_1.98-1.14 GenomeInfoDbData_1.2.10 ggrepel_0.9.5
[34] irlba_2.3.5.1 listenv_0.9.1 spatstat.utils_3.0-4
[37] goftest_1.2-3 RSpectra_0.16-1 spatstat.random_3.2-3
[40] fitdistrplus_1.1-11 parallelly_1.37.1 DelayedMatrixStats_1.22.6
[43] leiden_0.4.3.1 codetools_0.2-19 DelayedArray_0.26.7
[46] tidyselect_1.2.0 farver_2.1.1 viridis_0.6.5
[49] ScaledMatrix_1.8.1 spatstat.explore_3.2-6 jsonlite_1.8.8
[52] BiocNeighbors_1.18.0 ellipsis_0.3.2 progressr_0.14.0
[55] ggridges_0.5.6 survival_3.5-8 tools_4.3.0
[58] ica_1.0-3 Rcpp_1.0.12 glue_1.7.0
[61] gridExtra_2.3 xfun_0.42 withr_3.0.0
[64] fastmap_1.1.1 fansi_1.0.6 rsvd_1.0.5
[67] digest_0.6.34 timechange_0.3.0 R6_2.5.1
[70] mime_0.12 colorspace_2.1-0 scattermore_1.2
[73] tensor_1.5 spatstat.data_3.0-4 utf8_1.2.4
[76] generics_0.1.3 data.table_1.15.2 httr_1.4.7
[79] htmlwidgets_1.6.4 S4Arrays_1.0.6 uwot_0.1.16
[82] pkgconfig_2.0.3 gtable_0.3.4 lmtest_0.9-40
[85] XVector_0.40.0 htmltools_0.5.7 carData_3.0-5
[88] dotCall64_1.1-1 scales_1.3.0 png_0.1-8
[91] knitr_1.45 rstudioapi_0.15.0 tzdb_0.4.0
[94] nlme_3.1-164 zoo_1.8-12 KernSmooth_2.23-22
[97] vipor_0.4.7 parallel_4.3.0 miniUI_0.1.1.1
[100] pillar_1.9.0 grid_4.3.0 vctrs_0.6.5
[103] RANN_2.6.1 promises_1.2.1 BiocSingular_1.16.0
[106] car_3.1-2 beachmat_2.16.0 xtable_1.8-4
[109] cluster_2.1.6 beeswarm_0.4.0 evaluate_0.23
[112] cli_3.6.2 compiler_4.3.0 rlang_1.1.3
[115] crayon_1.5.2 future.apply_1.11.1 ggsignif_0.6.4
[118] labeling_0.4.3 ggbeeswarm_0.7.2 plyr_1.8.9
[121] stringi_1.8.3 BiocParallel_1.34.2 viridisLite_0.4.2
[124] deldir_2.0-4 munsell_0.5.0 lazyeval_0.2.2
[127] spatstat.geom_3.2-9 Matrix_1.6-5 RcppHNSW_0.6.0
[130] hms_1.1.3 patchwork_1.2.0 bit64_4.0.5
[133] sparseMatrixStats_1.12.2 future_1.33.1 shiny_1.8.0
[136] ROCR_1.0-11 igraph_2.0.2 broom_1.0.5
[139] bit_4.0.5 date()[1] "Thu Mar 14 00:22:46 2024"